All published articles of this journal are available on ScienceDirect.
Evaluation of Salivary KCNJ3 mRNA Levels in Breast Cancer: A Case–control Study and in silico Analysis
Abstract
Background:
Breast cancer (BC) is considered the most malignant and central cancer-related death among women worldwide. There is an essential need to discover new methods for developing noninvasive and low-cost diagnoses. The present study examines the expression of KCNJ3 which acts as a biomarker for detecting BC in the saliva of BC patients compared to controls.
Methods:
The mRNA expression level of KCNJ3 has been evaluated. Forty-three unstimulated whole saliva samples from BC patients and forty-three salivary samples from healthy controls were collected. The mRNA level was measured using quantitative real-time polymerase chain reaction (qRT-PCR). Furthermore, the protein-protein interaction network in which KCNJ3 is involved was obtained. In silico analysis was applied to predict the possible molecular mechanisms of KCNJ3 in BC development.
Results:
Differentially expressed KCNJ3 was statistically significant between BC patients and controls (p<0.001). The sensitivity and specificity of KCNJ3 mRNA in BC detection were 76.74% and 94.95%, respectively. Receiver operating characteristic (ROC) curve analysis of KCNJ3 mRNA revealed that Area under the curve (AUC) was 0.923 (95% Confidence Interval (CI): 0.866-0.979). AUCs of ROC curve analysis were 0.743 (95% CI: 0.536-0.951), 0.685 (95% CI: 0.445-0.925), and 0.583(95% CI: 0.343-0.823) for differentiation stage I from stage III, stage I to stage II and finally stage II from stage III, respectively. Furthermore, the GABAergic synapse signaling pathway was suggested as a potential pathway involved in BC development.
Conclusion:
Salivary levels of KCNJ3 could be considered a potential diagnostic biomarker with high sensitivity and specificity for BC detection.
1. INTRODUCTION
Breast cancer (BC) is considered the most malignant and primary cancer-related death in women worldwide [1]. Based on hormonal factors and molecular classification, BC is divided into three groups: estrogen receptor-positive (ER+), progesterone receptor-positive (PR+), and human epidermal growth factor receptor (HER2), which are the most common categories for the treatment and prediction of outcomes [2]. Genetically, mutations in the BRCA1 and BRCA2 genes have been reported in BC patients. In addition, mutations in other tumor suppressor genes, p53 and PTEN, have also been reported. Dysregulation of repair system components such as ATM, CHEK2, NBN, PALB2, and RAD50 can also be considered genetic risk factors for BC [3].
According to the World Health Organization, mammography is the best way to screen BC [4]. It has been introduced as an effective and, of course, a high-cost method. Recently, using salivary biomarkers has attracted attention in diagnosing diseases and cancers [5]. Research has been done to diagnose and prognosis using body fluids [6, 7]. Using saliva as a source of diagnostic biomarkers has advantages over blood. First, saliva is readily accessible. In addition, the collection of saliva is noninvasive, less expensive, and does not require trained personnel. Despite these advantages, not all biomarkers are detectable in saliva, primarily because of the difference in biomarkers’ concentrations in blood and saliva [8]. Several attempts have differentiated BC patients from healthy control regarding chemical, protein, metabolic, and transcriptomic biomarkers in saliva [9-11].
The growth and metastasis of cancer cells require a series of complex signals. These signals are transmitted into the cell by the surface receptors on the membrane. In the past, abnormal ion channel expression has been observed in many cancers [12, 13]. G-Protein-Coupled Inwardly Rectifying Potassium (GIRKs) is a group of k+ channels that connect membranes and extracellular signaling molecules [14]. These channels act as G proteins and regulate cell function by stimulating neurotransmitters and hormones. These channels comprise GRIK1 proteins encoded by the KCNJ3 gene. Increased expression of the KCNJ3 gene, which increases GRIK1 mRNA, is also associated with lung cancer, pancreatic adenocarcinoma, and breast cancer. Up-regulation of the KCNJ3 gene in breast tissue samples has been reported to cause down-regulation of p53 and up-regulation of estrogen receptor (ER). Patients with ER+ breast cancer and high KCNJ3 expression have a worse prognosis and lower survival than ER+ patients with low expression of this gene. Therefore, ER+ patients can be divided into high-risk and low groups based on their expression levels. Based on all this evidence, the study of salivary KCNJ3 gene expression in BC is valuable for validating this gene as an independent and prognostic biomarker for BC patients [13, 15].
Several studies have examined the effects of potassium channels and ions on various diseases in the saliva and the interaction effects of the disorders and potassium ions [ 16, 17]. Furthermore, calcium ions are known to play an essential role in the progression of BC [18, 19]. This study aimed to evaluate the salivary KCNJ3 expression level of BC patients and healthy controls. Furthermore, in combination with in silico analysis, this study examined the possible roles of KCNJ3 during BC development.
2. MATERIALS AND METHODS
2.1. Patient Enrollment and Samples
Forty-three BC women patients in Imam-Khomeini Hospital (Tehran, Iran), and forty-three women were referred to the dental clinic of Imam-Khomeini Hospital for routine oral examinations as controls were enrolled (Fig. 1). BC in patients was confirmed by histopathological and biopsy results. The demographic characteristics of the enrolled persons in the study are shown in Table 1.
| - | BC Patients (n=43) | Healthy Volunteer (n=43) |
|---|---|---|
| Age, years | 50.26±12.87 | 49.35±13.88 |
| Mean ± standard deviation (SD) | ||
| Range | 25-85 | 21-80 |
| Smoking, n (%) | 1 (2.3%) | 2 (4.6%) |
| Alcohol, n (%) | 0 (0%) | 0(0%) |
| Stages | N/A | |
| Stage I | 11 | |
| Stage II | 18 | |
| Stage III | 14 | |
| Hormone receptor status | N/A | |
| Estrogen receptor (ER+) | 36 | |
| Estrogen receptor (ER-) | 7 | |
| progesterone receptor (PR+) | 32 | |
| progesterone receptor (PR-) | 11 | |
| HER2 status | N/A | |
| Score 0, 1 (Negative) | 21 | |
| Score 2 (Borderline) | 11 | |
| Score 3 (Positive) | 11 | |
| Ki-67 labeling Index | N/A | |
| 0%-15% | 25 | |
| 15%-30% | 7 | |
| 30%-100% | 11 | |
| Subtype | N/A | |
| Invasive ductal carcinoma (IDC) | 37 | |
| Invasive lobular carcinoma (ILC) | 2 | |
| Ductal carcinoma in situ (DCIS) | 4 | |
| IDC/DCIS | 5 | |
| Triple-negative | 3 | N/A |
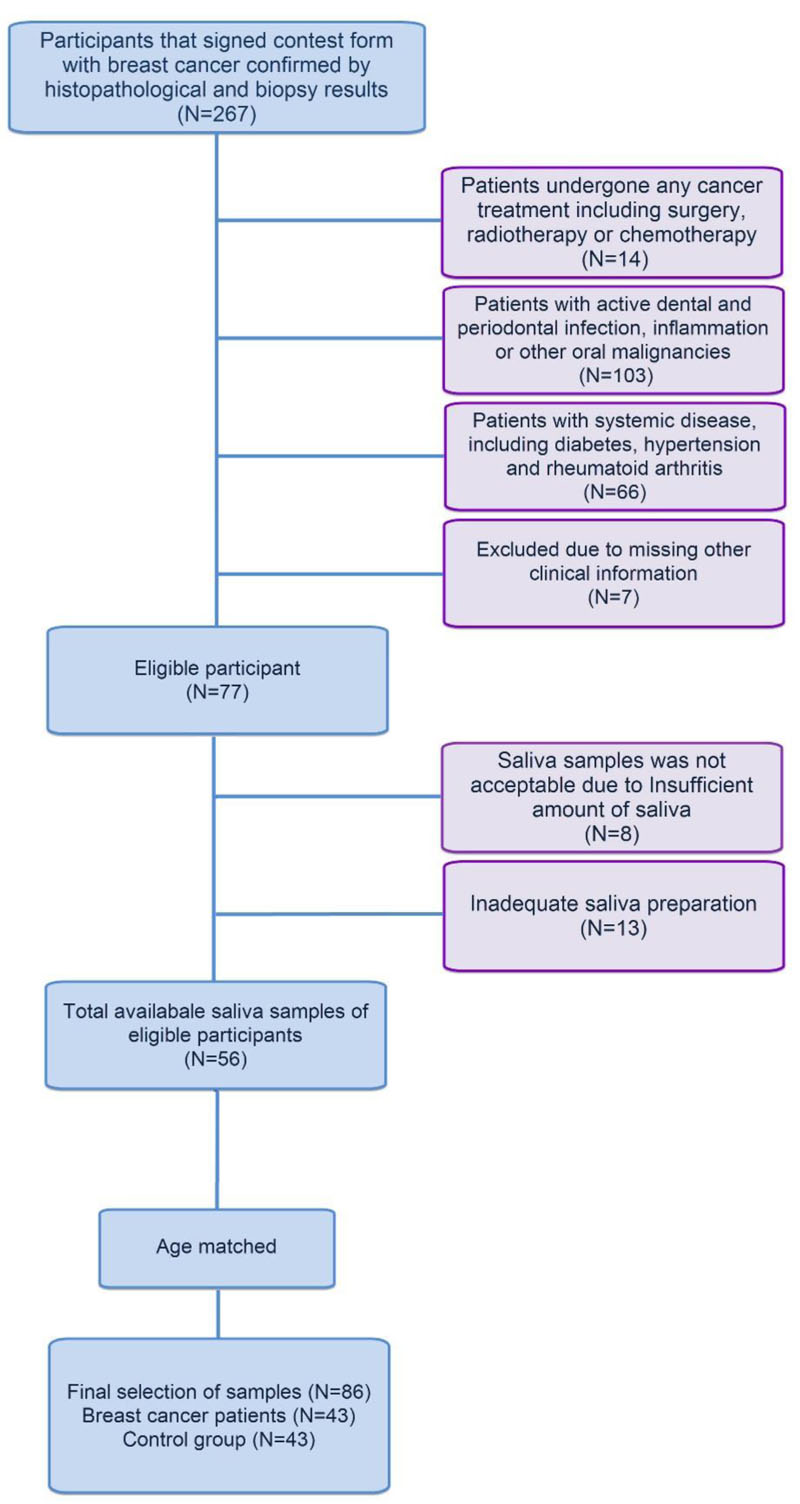
Besides, patients have undergone no surgery, radiotherapy, or chemotherapy, and they were free of any active dental and periodontal infection, inflammation, or other malignancies. The patient also had no systemic diseases, including diabetes, hypertension, and rheumatoid arthritis. The enrolled people in the control group also had no active mouth infections, inflammation, or other malignancies and systemic diseases. Subjects in the case and control groups were matched based on age. After obtaining signed informed consent from the participants, 10 ml of unstimulated whole saliva samples were collected by the spitting method. To prevent the probable effect of circadian rhythm, sampling was done between 8-10 AM. Saliva samples were stored in -80 oC refrigerator. For avoiding detection bias, data analysis was performed in the blinding method. The local Ethics Committee of Tehran University of Medical Sciences approved the study (Ethical code: IR.TUMS.DENTISTRY.REC.1398064).
2.2. RNA Isolation
Total RNA extraction from saliva was performed by using RiboEX (GeneAll, Seoul, South Korea) RNA Extraction kit according to the manufacturer's protocol. Moreover, the NanoDrop spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) was employed to evaluate the purity, quality, and integrity of the extracted RNA. After RNA isolation, the eluted samples were stored at -80 °C.
2.3. cDNA Synthesis And Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
cDNA synthesis was performed by BioFact kit (BioFact, Daejeon, South Korea), according to the manufacturer's protocol. For this purpose, the RNA samples were incubated at room temperature for 5 minutes, 10 minutes at 25 °C, 60 minutes at 42 °C and 10 minutes at 75 °C. For quantitative real-time polymerase chain reaction (qRT-PCR), the Ampliqon SYBR Green master mix (Stenhuggervej, Denmark) and KCJN3 specific primers were used (Table 2). All reactions were performed in triplicate. qRT-PCR was performed by Rotor-Gene Q (Qiagen, Germany) as follows; one cycle of 5 minutes at 95 °C, 40 cycles of 30 seconds at 95 °C, 30 seconds at 60 °C, 30 seconds at 72 °C, and one cycle of 5 minutes at 72 °C. The relative expression levels of miRNAs were calculated using the 2-ΔΔCt method relative to the actin gene (ACTB).
| Primer Sequences | Genes |
|---|---|
| 5'TGAGGGACGGAAAACTCACG3' | KCNJ3 Forward |
| 5'GGCTGGTTCTCAGACGAGTT3' | KCNJ3 Reverse |
| 5'GATCAAGATCATTGCTCCTCCTG3' | ACTB Forward |
| 5'CTAGAAGCATTTGCGGTGGAC3' | ACTB Reverse |
2.4. Statistical Analysis
SPSS software (version 22; SPSS Inc., Chicago, IL, USA) and GraphPad Prism 8.2.1 for Windows (GraphPad Software, San Diego, California) were applied to analyze the qRT-PCR results. Mann-Whitney test was applied to compare KCJN3 expression levels between case and control groups. Kruskal-Wallis test was used to analyze the expression level in various BC stages. Pearson’s test was applied to analyze the correlation between age and gene expression levels. Finally, receiver operating characteristic (ROC) curve analysis was done to evaluate the potential role of KCJN3 as a diagnostic biomarker in BC patients. All results are presented as mean ± standard deviation (SD), and p≤0.05 was considered statistically significant.
2.5. In Silico Analysis; Protein-protein Interaction (PPI) Network And Enrichment Of KCNJ3-relatEd Signaling Pathways
The string app in Cytoscape v3.7 [20] was employed to obtain the module in which KCNJ3 is involved (maximum number of interactors=0 and confidence score≥0.9). Next, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [21] was used to identify the signaling pathways in which the module is involved. For this purpose, the human genome was set as the background, and signaling pathways were enriched based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [22, 23].
3. RESULTS
None of the people in the case and control groups used alcohol, and only one person in the case group and two people in the control group smoked. There were also 11 patients in stage I, 18 patients in stage II, and 14 patients in stage III.
3.1. Expression Analysis of KCNJ3
Using qRT-PCR, the relative expression of KCNJ3 in 43 saliva samples of BC patients and 43 saliva samples of controls was assessed. Besides, the expression level of ACTB in all samples was obtained. Considering ACTB as a reference gene, the relative expression of KCNJ3 was determined. Here, we found that the relative expression of KCNJ3 was significantly up-regulated in BC patients compared to healthy controls (p<0.001) (Fig. 2).
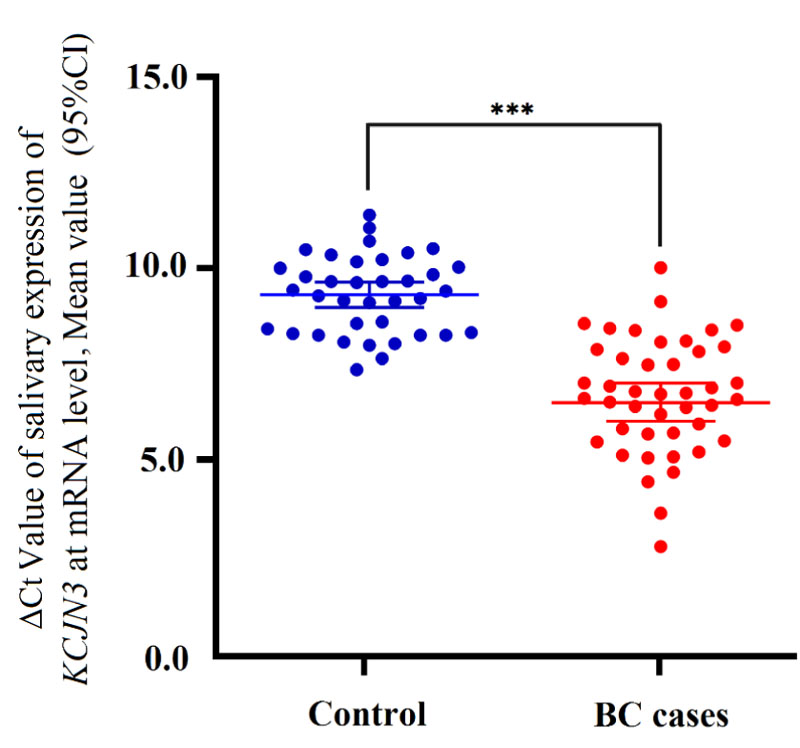
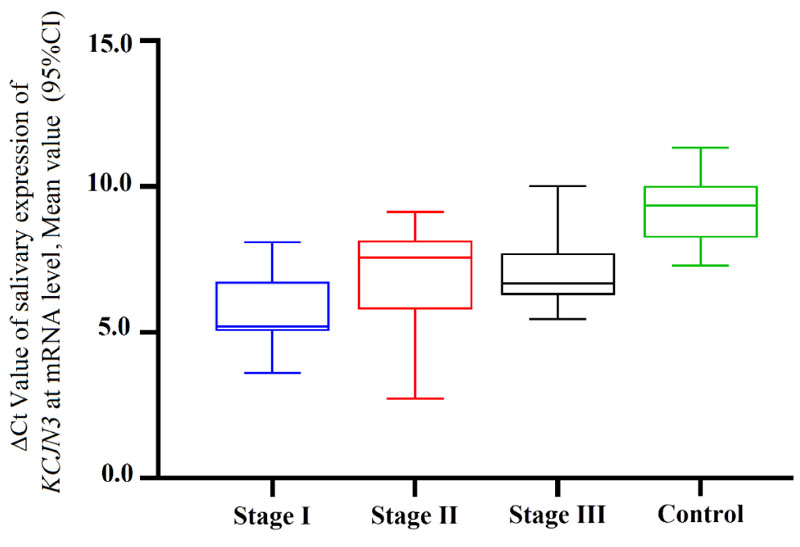
Furthermore, the differentially expressed KCNJ3 was not statistically significant among various BC stages (p=0.061) (Fig. 2B).
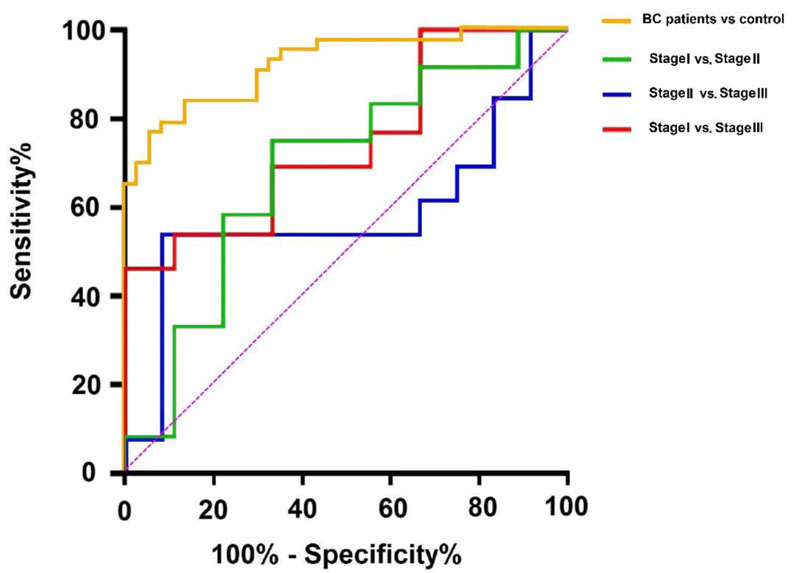
To determine the linear correlation between age and gene expression, the Pearson correlation analysis was performed. It was determined that there was no correlation between age and the salivary expression of KCNJ3 in BC patients (p:0.077). Considering the cut-off value of 7.900, the sensitivity and specificity of KCNJ3 mRNA in BC diagnosis were 76.7% and 94.59%, respectively. The area under the curve (AUC) of ROC curve was 0.923 (95%CI 0.866-0.979) (Fig. 3). Considering cut-off values of 4.389 for differentiation of stage I vs. III, 4.148 for stage I vs. II, and 3.885 for stage II vs. III, we revealed that the AUC of ROC curves were 0.743 (95% CI: 0.536-0.951), 0.685 (95%CI 0.445-0.925) and 0.583 (95%CI 0.343-0.823) for stage I vs. III, stage I vs. II and finally stage II vs. III, respectively.
The positive likelihood ratio (PLR) for salivary KCNJ3 was 14.20 (95% CI: 3.65-55.20) and the negative likelihood ratio (NLR) was 0.25 (95% CI: 0.14-0.43). PLR and NLR for discrimination of stage I vs. III were 2.077 (95% CI: 0.770-5.600), 0.461 (95% CI: 0.181-1.178). Meanwhile, PLR and NLR for differentiation of stage I vs. II: 2.250 (95% CI: 0.844-5.995), 0.375 (95% CI: 0.127-1.108) and PLR and NLR for diagnosis of stage II vs. III were 6.461 (95% CI: 0.926-45.094), 0.503 (95% CI: 0.273-0.928) respectively. ROC analysis results suggested that the KCNJ3 expression level could be considered a diagnostic biomarker when comparing BC patients with healthy controls (Fig. 4).
3.2. Signaling Pathway Analysis
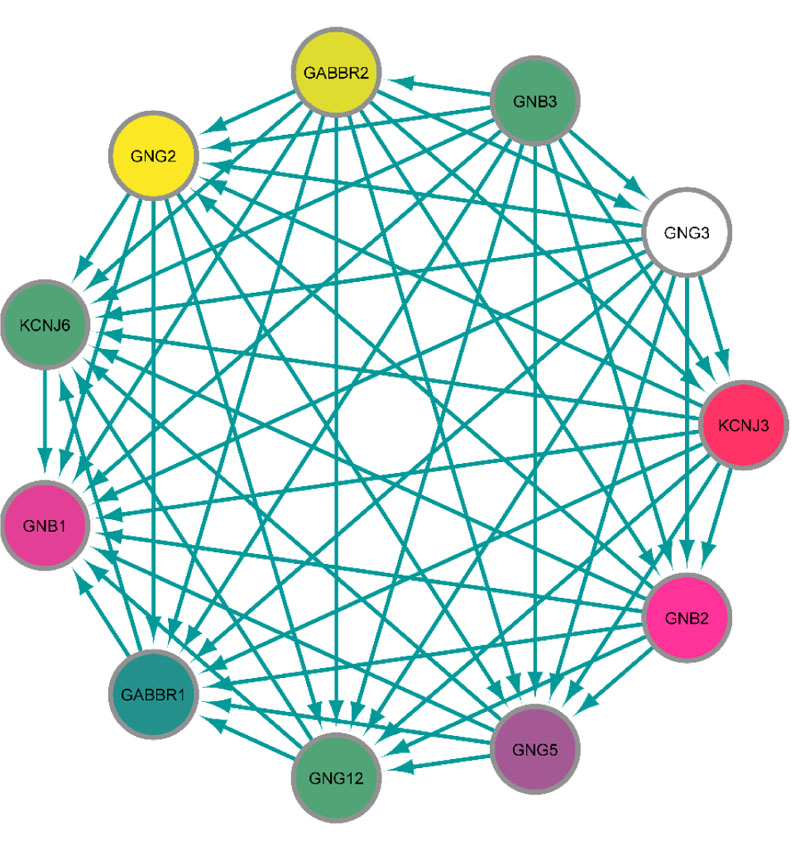
The protein-protein interaction (PPI) network in which KCNJ3 is involved was obtained using the string app in Cytoscape. Except for KCNJ3, ten proteins correlate with each other in the main module with a high confidence score (≥0.9) (Fig. 5).
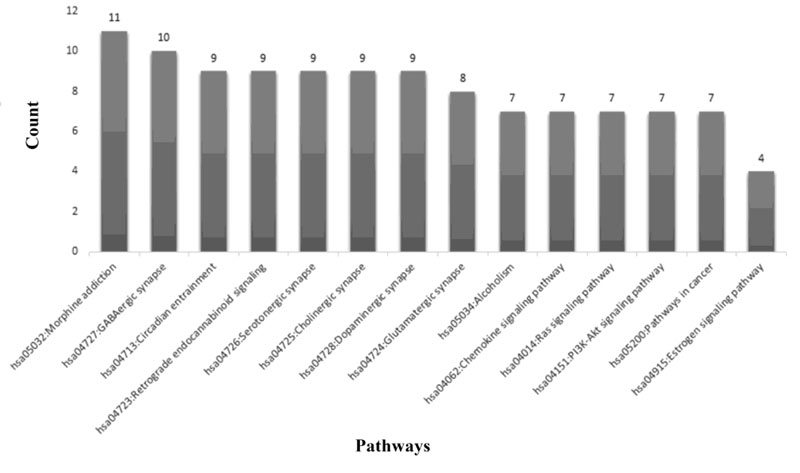
Furthermore, the DAVID database was applied to enrich module-related signaling pathways. As is evident in Fig. (5), Morphine addiction (p=9.89E-20), GABAergic synapse (p=4.31E-17), and circadian entrainment (p=4.32E-14) are the top enriched pathways (Fig. 6).
4. DISCUSSION
To date, several studies have been performed to evaluate salivary biomarkers in patients with BC. There are some benefits to using saliva to consider as biomarkers in malignancies. The most crucial advantage of saliva is the noninvasive sampling process and it is more accessible than serum or tissue sampling [24]. Moreover, since saliva sampling does not require special tools and equipment, screening efforts for anomalies like cancer are better than serum [25, 26]. Zhang et al. showed that the origin of salivary biomarkers in BC is probably due to the common embryonic origin of salivary gland tissue and breast tissue [9]. Meaning that many of the markers produced by breast tissue are also produced and secreted by the salivary glands. Therefore, common biomarkers have been observed and found in malignancies of both tissues [27]. Zhang et al. identified CSTA, TPT1, S100A8, MDM4, MD6, H6PD, GRIK1, and GRM1 mRNAs in BC patients' saliva. According to this study, eight mRNAs have an acceptable potential for BC diagnosis with 83% sensitivity and 97% specificity [9].
For the first time, the mRNA expression level of KCNJ3 gene in BC patients' saliva relative to healthy controls was evaluated. The results showed significant up-regulation of KCNJ3 mRNA expression in BC patients' saliva samples compared with the healthy control group. Previous studies have shown that increased expression of KCNJ3 mRNA is associated with BC metastasis to lymph nodes [28]. This gene is also up-regulated in primary and invasive BC [29]. In a study performed by Kammerer et al. on more than 1000 patients with BC showed that the KCNJ3 gene was up-regulated in breast tissues compared to normal tissues [28]. Rezania et al. showed that the expression level of KCNJ3 in patients with early-stage BC was significantly up-regulated compared to healthy individuals [15], which is consistent with the present study. Based on the ROC curve analysis, it was found that KCNJ3 expression level could be considered a diagnostic biomarker in BC patients to evaluate this biomarker's sensitivity and specificity in the diagnosis of BC. Relying on its relatively high sensitivity (76.7%), patients can be easily identified as healthy individuals and its high specificity (94.59%) minimizes false positives. Therefore, this gene's salivary value could be considered a potential biomarker for the diagnosis of BC. Positive likelihood ratio higher than 10 and a negative likelihood ratio less than 0.1 results in an acceptable potential of salivary KCNJ3 in BC diagnosis. KCNJ3 expression levels were assessed in various disease stages; no statistically significant KCNJ3 expression alterations were found among the various stages. Therefore, it could be claimed that from the beginning to the final stages, KCNJ3 expression level is increasing, so it could be considered a diagnostic biomarker for early-stage BC. However, considering the salivary expression levels are not significantly different in the disease stages, this could be due to the study's small sample size. Based on the study by Lopez-Jornet et al., it is suggested that CA125 can be a potential diagnostic salivary biomarker besides KCNJ3 mRNA level with acceptable sensitivity and specificity [30]. Finally, the PPI network analysis determined the leading KCNJ3 gene network. Ten proteins were determined, including GNG12, GNG5, GNG3, GNB2, GNB3, GABBR2, GNG2, GABBR1, KCNJ6, and GNB1, high correlate with the KCNJ3 protein. Moreover, using in silico analysis, the signaling pathway in which KCNJ3 is involved was enriched (Fig. 5). Gabaergic synapse signaling was enriched as one of the related pathways. γ-Aminobutyric Acid (GABA) is an inhibitory neurotransmitter, especially in the nervous system. However, the existence of GABA and GABA receptors in various tissues, including lung and oviduct with crucial roles, has been determined. Furthermore, it has been reported that GABA activity is increased in various types of cancers such as colon, gastric, and breast cancer and may affect the proliferation, growth, and metastasis of cancer cells [31, 32]. Zhang et al. proved that the GABAerigc signaling system is present in BC cells. GABAB receptor signaling promotes BC invasion in vitro and migration in vitro and in vivo, mediated by phosphorylation of ERK1/2 and, subsequently, up-regulation of matrix metalloproteinases-2 [33]. However, further studies are needed to clarify the effect of GABA on BC cell proliferation. Estrogen signaling is another pathway in which KCNJ3 gene is involved. Estrogen receptors belong to the transcription factor family, a key regulator of involved genes in cell proliferation and migration. It has been reported that dysregulation and aberrant expression of ER signaling components lead to BC development and invasion [34]. For future studies, assessing the potential of KCNJ3 mRNA levels for breast cancer screening and the possibility of KCNJ3 protein for breast cancer diagnosis and targeted therapy is suggested.
CONCLUSION
This research was conducted to compare the levels of KCNJ3 salivary expression in BC patients with healthy individuals. Differentiation criteria based on the salivary expression levels of KCNJ3 were proposed to distinguish BC patients from healthy controls, using statistical and in silico analyses. K CNJ3 expression at mRNA level in saliva could be a potential biomarker to detect BC from healthy control, especially in the primary stages of BC. Moreover, regarding the enriched signaling pathways such as GABAergic synapse signaling and estrogen signaling, further studies are needed to elucidate the exact role of KCNJ3 in the development of BC.
LIST OF ABBREVIATIONS
| AUC | = Area under the curve |
| BC | = Breast cancer |
| CI | = Confidence interval |
| DCIS | = Ductal carcinoma in situ |
| ER | = Estrogen receptor |
| ER+ | = Estrogen receptor-positive |
| GABA | = γ-Aminobutyric Acid |
| HER2 | = Human epidermal growth factor receptor |
| IDC | = Invasive ductal carcinoma |
| ILC | = Invasive lobular carcinoma |
| KEGG | = Kyoto Encyclopedia of Genes and Genomes |
| NLR | = Negative likelihood ratio |
| PLR | = Positive likelihood ratio |
| PR+ | = Progesterone receptor-positive |
| PPI | = Protein-protein interaction |
| qRT-PCR | = Quantitative real-time polymerase chain reaction |
| ROC | = Receiver operating characteristic |
| SD | = Standard deviation |
AUTHORS' CONTRIBUTIONS
MK and MJ conceived the study idea. MK, MJ, SM, and RM created the study protocol, led the data collection, and wrote the original draft. MK, MJ, and RM contributed to the data analysis/interpretation and preparation of the manuscript. MK and SM led the writing- review and editing. MK and SK interpreted the results. All authors have read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by Tehran University of Medical Science Ethical Committee (ethical code: IR.TUMS.DENTISTRY.REC.1398064).
HUMAN AND ANIMAL RIGHTS
No animals were used for studies that are the base of this research. All the human procedure were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
After describing the study objectives, written informed consent was obtained from all participants.
STANDARD OF REPORTING
STROBE guidelines were followed.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study are available from the corresponding author [S.M] on reasonable request.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests.
FUNDING
There is no source of financial support or funding.
ACKNOWLEDGEMENTS
The authors would like to appreciate all participants who contributed and provided data to make this study possible.


