The Role of Periodontopathogens and Oral Microbiome in the Progression of Oral Cancer. A Review
Abstract
Chronic periodontal disease and oral bacteria dysbiosis can lead to the accumulation of genetic mutations that eventually stimulate Oral Squamous Cell Cancer (OSCC). The annual incidence of OSCC is increasing significantly, and almost half of the cases are diagnosed in an advanced stage. Worldwide there are more than 380,000 new cases diagnosed every year, and a topic of extensive research in the last few years is the alteration of oral bacteria, their compositional changes and microbiome. This review aims to establish the relationship between bacterial dysbiosis and OSCC. Several bacteria implicated in periodontal disease, including Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, and some Streptococcus species, promote angiogenesis, cell proliferation, and alteration in the host defense process; these same bacteria have been present in different stages of OSCC. Our review showed that genes involved in bacterial chemotaxis, the lipopolysaccharide (LPS) of the cell wall membrane of gram negatives bacteria, were significantly increased in patients with OSCC. Additionally, some bacterial diversity, particularly with Firmicutes, and Actinobacteria species, has been identified in pre-cancerous stage samples. This review suggests the importance of an early diagnosis and more comprehensive periodontal therapy for patients by the dental care professional.
1. INTRODUCTION
Oral bacteria dysbiosis is known to act as a precursor for diseases such as periodontitis [1, 2]. This pathologic process, which involves chronic inflammatory responses that increase the possibility for tooth loss, can also affect systemic health because its accumulative effect increases the patients’ risk for neoplastic processes [3-5]. Head and neck cancer is the sixth most common human cancer [6], representing 3% of all types of cancer. Lesions of the oral cavity represent 48% of head and neck cases, with 90% of oral cavity lesions being squamous cell carcinoma (OSCC), which is one of the most common causes of death among all oral cancers [7]. Prognosis is always dependent on age, lymph node involvement, tumor size and location [7]. The development of OSCC is statistically linked to several classical risk factors, including tobacco, alcohol abuse, Human Papillomavirus (HPV), poor hygiene and predisposing conditions, which induce pro-oncogenic and epigenetic alterations [8-10]. However, other factors, including oral inflammatory diseases, infections, and bacterial dysbiosis, are recognized as risk factors for cancer development [11, 12].
Evidence has demonstrated that a combination of risk factors such as smoking, alcohol abused, and poor oral hygiene increases the risk of oral cancer by inducing chronic inflammation and infection; these risk factor modify the local microbiota and host interactions, altering homeostasis of the oral environment [11, 13-15]. On the other hand, the development of OSCC in subjects who had never used tobacco or alcohol suggests that chronic periodontitis might be an independent risk factors for cancer development [16]. The causal relationship between specific oral bacterial infections and the development of OSCC, has been supported by the presence of some oral periodontopathogens such as Porphyromonas gingivalis (P. gingivalis), or Fusobacterium nucleatum (F. nucleatum); these periodontopathogens are known to stimulate different cytokines which facilitate cell proliferation, mutagenesis, apoptosis inhibition, oncogene activation, and angiogenesis [15, 17-20].
The most abundant microbiota is found in the gut, followed by the oral cavity; Oral Cancer (OC) has been associated with oral and gut microbiota dysbiosis [11, 12]. This complex interaction also lays the foundation of the crosstalk between oral microbiota and gut microbiota [21, 22]. Oral bacteria can be transported into the gut and change the gutmicrobiota affecting the immune defense system [23-25], especially in patients diagnosed with chronic periodontitis, comorbidities, systemic diseases and immunocompromised individuals [3, 26, 27]. Therefore, oral microbiota is emerging as a complex factor influencing oral microenvironments, oral diseases and OSCC, but also affecting the gut microbiota in both direct and indirect ways [11, 21]. The most representative bacteria associated with OSCC are F. nucleatum, P. gingivalis, and Prevotella intermedia [17, 28-31]. Furthermore, other bacterial species, such as Actinomyces, Clostridium, Enterobacteriaceae, Fusobacterium, Haemophilus, Porphyromonas, Prevotella, Streptococcus spp. and Veillonella have been associated with pre-cancerous lesions and OSCC [31, 32]. However, the cellular and molecular events that these bacteria promote in the host are not totally understood.
To understand the influence in the oral carcinogenesis process associated with oral dysbiosis, a narrative review was performed to summarize the microbiological, cellular, molecular and clinical aspects from in-vitro and clinical studies, thus providing an updated and structured overview of the oral microbiota and oral cancer relationship.
2. THE CLINICAL ASSOCIATION BETWEEN PERIODONTAL DISEASES AND ORAL CANCER
OSCC constitutes almost 90% of all oral cancers and is the leading cause of death among all oral cancers [33]. OSCC originates from the oral epithelial mucosa with more than 380,000 cases reported annually worldwide and more than 175,000 recorded deaths worldwide in 2018 [34]. More than 50% of the cases are diagnosed in advanced stages [35]. It occurs more frequently in men, with a higher incidence in South Asia, Western and Eastern Europe [36]. In Colombia, more than 140 to 160 new cases of OSCC occur annually, with 50% occurring in the tongue, and it is threefold more frequent in men than in women [37].
Chronic Periodontal Disease (CPD) is highly prevalent in adults, and its severity increases with age. The relationship between CPD and oral cancer has been examined for some years. Numerous case-controlled studies have addressed the role of CPD in head and neck cancer, and several cohort studies have examined its association with other types of cancers [38]. PubMed, Embase, Science direct, and Scopus searches were performed using keywords such as “oral bacteria” OR “oral cancer” AND “dysbiosis” OR “chronic periodontal disease” AND “oral microbiome”. This review included studies only published in English.
To date, six meta-analyses or reviews have been published about the associations between periodontal disease and/or tooth loss, and the risk of head and neck cancers [38-44] (no randomized controlled trials exist, with relevant exposure categories and outcomes). In fact, given the importance of smoking as a potential promoter for OSCC, only papers with estimated adjustment for smoking were considered [38].
Only one cohort study reports smoking-adjusted associations for periodontal disease and head and neck cancer, which showed low risk, and was not statistically significant (HR = 1.15, 95% CI: 0.73, 1.81) [1]. In the same cohort, after the analyses were limited to non-smokers, a strong association was observed for periodontitis, oropharyngeal and esophageal cancers combined (HR = 2.25, 95% CI: 1.30, 3.90) [44]. Regarding the number of teeth, many studies have been conducted to evaluate general oral health concerning to head, neck and esophageal cancers. However, most of these reports are case-controlled. studies with several limitations, including large clinical heterogeneity and incompatible measures of periodontal disease; this has led to the impossibility of conducting accurate and feasible meta-analysis studies. Therefore, cohort studies of periodontitis using standardization of measurements are required to overcome statistical heterogeneity, publication bias and not unified adjusted factors. Although recent studies with improved measurements of periodontitis support strong positive associations for periodontitis and oral cancer risk [45], well-designed cohort studies are still needed to determine a more accurate relationship between these entities [38, 39].
2.1. Oral and Subgingival Dysbiosis in Periodontitis and Cancer
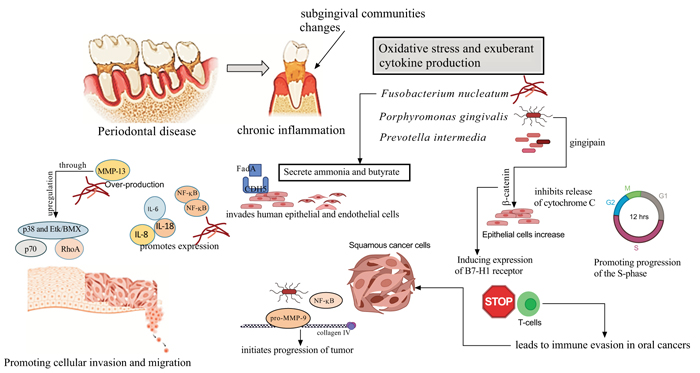
The onset of oral diseases as CPD involves significant changes in the composition of subgingival bacterial communities. Chronic inflammation and the anaerobic nature of the periodontal pocket results in gradual progression to subgingival dysbiosis, which is significantly promoted by keystone pathogens as P. gingivalis. In a murine study, it has been established that P. gingivalis- induced periodontitis requires the presence of the commensal microbiota because P. gingivalis alone is unable to cause periodontitis despite colonizing this host [40-46]. In addition, P. gingivalis is also influenced by the inflammatory environment orchestrated by pathobiont, such as F. nucleatum, prevotellas spp, and even commensal bacteria such as Streptococcusgordonii, Firmicutes and Actinobacteria [47-49].
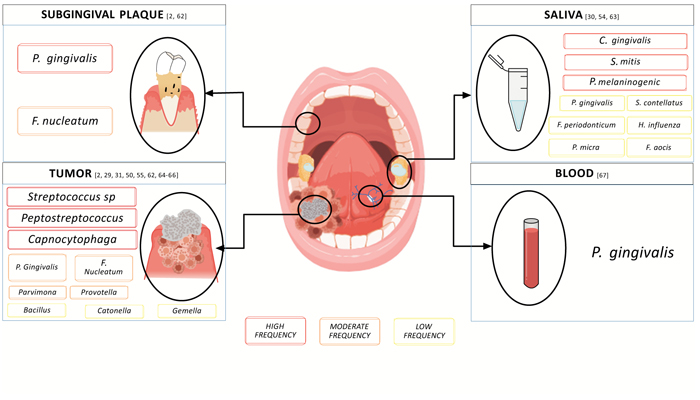
Significant shifts in the composition of the oral microbiome have been observed in oral cancers [50-57]. Several oral commensal bacteria, and keystone pathogens are correlated with OSCC and have been isolated from tumoral microenvironments (Table 1) [17, 18, 56, 58]. In this regard, three key microorganisms have been identified as being involved in subgingival dysbiosis, F. nucleatum, P. gingivalis, and Prevotella intermedia which are also considered the most representative types of bacteria in OSCC [17, 28-31, 59-63], (Fig. 1); An interesting feature is that P. gingivalis appears to occur in the early stage of OSCC 53, and F. nucleatum has also been identified as a key player in non-oral cancers (gastric, digestive, colorectal and pancreatic cancer) [64-67].
Table 1.
| Cancer (Subjects) | Bacteria Detected | Method | Site Isolation | Ref | Country, Year |
|---|---|---|---|---|---|
| OSCC (46) | Streptococcus anginosus | PCR | Intra tumoral & Dental Plaque | [65] | 2005 |
| OSCC (45) |
Capnocytophaga gingivalis, Prevotella melaninogenica Streptococcus mitis, |
DNA-DNA hybridization. | Saliva | [30] | 2005 |
| OSCC (125) | Bacillus, Enterococcus, Parvimonas, Peptostreptococcus, | 16S rRNA sequencing | Saliva | [66] | Taiwan, 2017 |
| OSCC (10) | Streptococcus sp., Peptostreptococcus, Gemella spp, Johnsonella ignava | 16S rRNA sequencing | Intra tumoral | [67] | USA 2012 |
| OSCC(5) | Streptococcus and Rothia | 16S rRNA sequencing | Intra tumoral | [54] | USA 2014 |
| OSCC (10) | P. gingivalis | immunohistochemical staining | Intra tumoral | [29] | USA 2011 |
| OSCC (30) | Streptococcus spp | Bacterial culture | Lymph nodes | [60] | Japan 1999 |
| OSCC (6) | P. gingivalis and F. nucleatum | 16S rRNA sequencing | Intra tumoral and subgingival plaque | [2] | Germany 2019 |
| OSCC (60) | T. denticula | Immunohistochemical staining | Intratumoral | [61] | Finland 2018 |
| OSCC (197) | Fusobacterium periodonticum, Parvimonas micra, Streptococcus constellatus, Haemophilus influenza, and Filifactor | 16S rRNA sequencing | Saliva | [53] | Taiwan 2018 |
| OSCC (40) | Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella and Parvimona | 16S rRNA sequencing | Intra tumoral | [49] | China 2017 |
| OSCC (7852) | P. gingivalis | Serum IgG antibody | Blood | [62] | USA 2012 |
| OSCC (21) | Veillonella, Fusobacterium, Prevotella, Porphyromonas, Actinomyces and Clostridium (anaerobes), and Haemophilus, Enterobacteriaceae and Streptococcus spp. | Bacterial culture | Intra tumoral | [31] | Hungary 1998 |
Composition of oral microbiome obtained from samples of saliva, blood, tumor, and subgingival plaque in patients with OSCC. We highlight P. and F. nucleatum as the main ones (Fig. 1).
3. ORAL MICROBIOME AND ITS RELATIONSHIP WITH ORAL CANCER STAGES
In terms of the composition of the oral microbiota, Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, and Actinobacteria were the five dominant phylum in the oral cavity. Six families (Prevotellaceae, Fusobacteriaceae, Flavobacteriaceae, Lachnospiraceae, Peptostreptococcaceae, and Campylobacteraceae) and 13 genera, including Fusobacterium, Alloprevotella, and Porphyromonas, were increased in OSCC. Species like F. nucleatum, P. intermedia, Aggregatibacter segnis, Peptostreptococcus stomatis, and Catonella morbi, showed a significant increase in oral cancer lesions [49-52]. Interestingly, Streptococcus, along with Actinobacteria (early oral colonizers), decrease the proinflammatory response orchestrated by F. nucleatum in the epithelial cells. Moreover, low levels of Firmicutes and Actinobacteria also promote a pro cancer environment [59], Noteworthy, F. nucleatum increases substantially in OSCC cases [50, 68].
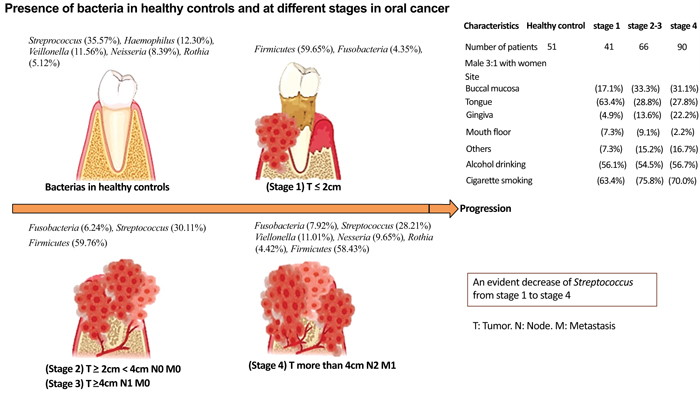
Different bacteria have been identified at different oral cancer stages. The clinical cancer stages of the 7th AJCC [69], and the oral microbiome present at each stage are depicted in Fig. (2). Yang C, et al. 2018, analyzed the microbiota in 248 samples from oral rinses that were collected within 1-3 weeks after the disease diagnosis in 51 healthy individuals and 197 OSCC patients at different stages, 53 using 16S rRNA V3V4 amplicon sequencing [53]. Fusobacteria increased significantly with the progression of oral cancer in comparison to healthy controls (2.98%) compared to OSCC stage 1 (4.35% p = 0.0015), stages 2 and 3 (6.24%, p < 0.0001), and compared to stage 4 (7.92% p < 0.0001), Fusobacteria was the most abundant in stage 4 [53]. As oral cancer progresses, Bacteroidetes and Actinobacteria decrease from stage 1 to stage 4 67. The main bacteria in OSCC stage 4 were: Streptococcus (28.21%), Veillonella (11.01%), Neisseria (9.65%), Haemophilus (9.37%), and Rothia (4.42%). The genera that increased the most was (Fusobacterium) (p < 0.0001), and the genera that decreased were: Streptococcus, Haemophilus, Porphyromonas, and Actinomyces (p < 0.0001) [53]. Firmicutes were the most dominant phylum in the oral rinse samples; being found in 58.40% of healthy patients, 59.65% in OSCC stage 1 patients, 59.76% in stage 2 and 3 patients, and 58.43% in OSCC stage 4.53 (Fig. 2).
4. THE ROLE OF THE ORAL MICROBIOME ON CARCINOGENESIS ASSOCIATED WITH ALCOHOL OR TOBACCO
Tobacco and alcohol are the main etiological factors in the development of OSCC [70-72]. Tobacco contains several carcinogenic molecules, especially polycyclic hydrocarbon, nitrosamines such as N'-nitrosonornicotine (NNN) and nitrosamine 4- (methylnitrosamine) -1 (-3-pyridyl) -1-butanone (NNK) [73]. The aforementioned molecules are known to be involved in DNA damage, interfering with replication, transcription and DNA repair. The nitrosamines induce chronic inflammatory processes due to the impairment of the defense mechanisms of the host and its microbiome [73-76].
Tobacco also promotes alterations in the oral microbiota, causing a decrease in bacteria such as Granulicatella, Staphylococcus, Veillonella, Capnocytophaga and an even greater decrease in Peptostreptococcus [77-80]; furthermore, species like Neisseria are not only inhibited by tobacco but also by alcohol consumption [78].
Tobacco use has been associated with the alteration of the surfaces of oral epithelial cells, allowing the growth of certain pathogenic bacteria [81, 82]. Showing significant increases of Prevotella parvimonas, Fusobacterium, Campylobacter, Bacteroides, Dialister, Treponema spp and Porphyromonasin, the oral microbiota have been identified with tobacco use [79, 80, 83]. Tobacco consumption affects bacterial growth in several ways, altering the cytotoxic effects on some bacteria [84], impairing saliva-bacterial interaction [85], and producing ethanol as a substrate for bacterial metabolism [86].
Acetaldehyde, the first metabolite of ethanol, is derived from alcohol consumption; its presence is associated with cellular damage, affecting the DNA repair process and has the potential to produce mutations in the DNA. Thus, acetaldehyde is considered a mutagenic and carcinogenic element. It has been associated with liver and upper aerodigestive tract cancer [85, 87].
In contrast alcohol dehydrogenase 2 (ALDH2) is an enzyme that decontaminates acetaldehyde products, but 2(ALDH2) polymorphisms may modify the association between alcohol use and pathogenic periodontal bacteria, as indicated by a higher percentage of pathogenic periodontal bacteria in saliva [88]. Some bacteria, such as Neisseria, Streptococcus and Actinomyces, reportedly have the ability to produce acetaldehyde [87, 89], although this is in contrast with a study by Thomas and Yokohama, which reported a low capacity for acetaldehyde production from these bacteria [78, 90]. Other bacterial species such as S. gordonii, S. mitis, S. oralis, S. salivarius, S. sanguinis R. mucilaginosa, P. histicola, S. salivarius and R. mucilaginosa, also have the capacity to produce acetaldehyde [89, 91, 92]. It has also been demonstrated that the alteration of the oral microbiome with alcohol, along with poor hygiene, enrich bacterial species such as Aggregatibacter, Actinomyces, Kingella, Leptotrichia, Cardiobacterium, Bacteroidales and Prevotella; while at the same time decreasing bacterial families such as Lactobacillales, Bacteroidetes, Firmicutes and Peptostreptococcaceae [92].
4.1. Molecular Mechanisms Implicated in the Pathogenesis of Oral Cancer
OSCC is a multiple-step molecular processes that develops from an individual genetic predisposition and/or exposure to external carcinogens. Some bacteria appear to be more prone to produce such changes. TP53 is a tumor suppressor gene that prevents carcinogenesis by activating G1 cell cycle arrest and promoting a p53 product [93], which induces apoptosis. However, 40–70% of oral cancers have mutations in the TP53 gene, with more than 90% of these mutations between exons 5 and 8 of TP [53, 93, 94]. These mutations can be separated into 2 categories: 1) disruptive, promoting an alteration in the DNA binding domains or damaging the p53 gene, with a subsequent failure of apoptosis and cell cycle arrest [95], and 2) non-disruptive, which only partially affects p53 function [96], producing deficient DNA repair, facilitating genomic instability, cellular proliferation, invasion, migration and dysregulating metabolism, ultimately leading to therapeutic resistance of cancer cells [97]. Another process is the loss of chromosomal region 9p21, which occurs in 70-80% of dysplastic lesions of the oral mucosa, being an early event in oral cancer [98]. The p16 tumor suppressor gene binds to the cyclin-dependent kinases CDK4 and CDK6, inhibiting cellular proliferation and avoiding synthesis phase entry in the cell cycle [99]. Expression of p16 is lost in 83% of oral cancers and 60% of premalignant lesions, heralding a poor prognosis [99]. On the other hand, Retinoblastoma 1 (RB1) is a tumor suppressor gene belonging to the RB family, which regulates the cell cycle at the G1-S checkpoint [100]. This gene is located on the long arm of the chromosome [16], an area with almost 17% of loss of heterozygosity in oral cancer [100, 101]. When hypophosphorylated it inhibits E2F activation, whereas its hyperphosphorylation by cyclin-dependent kinase complexes leads to pRb inactivation, releasing E2F activating target genes (c-MYC, n-MYC, CDC-2, p21WAF-1, cyclin A, c-MYB, and EGFR) involved in DNA synthesis, cell cycle regulation and cell growth [102], This process can be altered by HPV E7 oncogenes, dysregulating E2F1 and promoting inhibition of apoptosis, and the p53 mediated pathway, facilitating cancer cell progression [103]. Epidermal growth factor receptor (EGFR/ErbB1/HER1) is a proto-oncogene that belongs to the tyrosine kinase receptor family [104], Somatic mutations in EGFR are reported in over 90% of HNSCC and are linked with the inhibition of apoptosis in tumor cells, increased cell growth, angiogenesis and metastasis with poor prognosis [105]. Almost 3% of oral cancers have mutations in the tyrosine kinase domain of EGFR [ 106]. The Bcl-2 family comprises at least 15 proteins with either anti-apoptotic (Bcl-2, Bcl-XL) or pro-apoptotic effects (Bax, Bak). Alterations in the ratio of pro-apoptotic to antiapoptotic proteins promote tumor progression and can lead to oncologic therapy resistance [107]. Rat sarcoma virus (RAS) is a protooncogene, that is frequently mutated in oral cancer and the mutations occur mostly in H-Ras [108], being more common in smokers, even in some ethnic particular groups [109]. PTEN/mTOR/AKT/PI3K pathway appears to be a repeatedly dysregulated pathway in oral cancer, inducing some signaling pathways that suppress autophagy, thus reducing cell death [110]. These disorganized events result in activation of several intracellular messenger signaling systems, with an increased production of growth factors, along with evading inhibitory signals, leading to autonomous growth of tumor cells.
5. CELLULAR AND MOLECULAR PRINCIPLES OF MICROBIAL CARCINOGENESIS FOR PORPHYROMONAS GINGIVALIS
In primary cultures, P. gingivalis inhibited apoptosis by up-regulating the anti-apoptotic molecule Bcl-2 and down-regulating the pro-apoptotic Bax protein [111], thus promoting survival of primary gingival epithelial cells through activation of the pPI3K/Akt pathway [112], also improved the levels of PI3K and phosphoinositide-dependent protein-serine kinase 1 (PDK1) activated by Akt, promoting cell proliferation [113]. Furthermore, P. gingivalis aids gingival epithelial cells (GEP) progression through the G1 phase with less control, and more speed by down-regulating kinases such as Chk2, CK1delta, and CK1epsilon [114]. The suppression of the proapoptotic activity of Bax (Bcl-2-associated death promoter), results in blockage of the apoptosis effector cytochrome C [115], leading down-stream inhibition of caspase-9 and the executioner caspase-3 [115, 116]. Hoppe et al. found an increase in tumor cell proliferation with P. gingivalis dysregulates α-defensins [117]. P. gingivalis dysregulates some genes that intervene at the proinflammatory pathway of NF-κB with members of the MAPK family (like MAPK14 (p38), MAPK8 (JNK1), and NFKB1 (p50), which could promote cancer progression [118]. P. gingivalis activates a nucleoside diphosphate kinase (NDK), inhibiting apoptosis through purinergic receptor P2X7 in GEP [119]; NDK might also interfere with an anticancer immune response mediated by ATP activation of P2X7 receptors on dendritic cells [18, 120]. In this regard, the regulation of the cell cycle by miR-203 may affect the pathological expression in some carcinomas [121]. In addition, co-infection with P. gingivalis and F. nucleatum stimulates progression of chemically induced oral cancer in a murine model, via activation of the IL6/STAT3 axis; the suppression of mitochondrial-dependent apoptosis may be an important stratagem for the survival of P. gingivalis in periodontal tissues and a key feature of its pathogenicity [122].
6. CELLULAR AND MOLECULAR PRINCIPLES OF MICROBIAL CARCINOGENESIS FOR FUSOBACTERIUM NUCLEATUM
F. nucleatum LPS contain heptose, and2-keto-3-deoxyoctonate which may inhibit the intrinsic apoptotic pathway of oral epithelial cells [20]. Moreover, F. nucleatum enhance a pro-inflammatory response NF-kB gene activation and mitogen-activated protein kinase (p38). Accordingly, it drives the production of inflammatory cytokines, such as IL-1α, IL-1β, IL-6, IL-8, and MMPs, specially MMP13 (collagenase 3) [121-124]. In vitro studies have shown that F. nucleatum triggers Toll-Like Receptor (TLR) activation, stimulating IL-6 production, and activating STAT3, which induced cancer cells to grow by the cyclin D1 effector [123-125]. Interestingly, IL-6 was reported to induce vascular endothelial growth factor (VEGF) production in OSCC [126]. Thus, both P. gingivalis and F. nucleatum promote resistance to apoptosis by the activation of TLR2, which was seen more frequently in OSCC tissues than in normal tissues [125, 126]. F. nucleatum promotes cellular migration through stimulation of Etk/BMX, S6 kinase p70, and RhoA kinase [121, 124]. On the other hand, F. nucleatum promoted E-cadherin-expressing colorectal cancer cell (CRC) proliferation by modulating the E-cadherin/β-catenin signaling pathway or by activating TLR4 signaling to NF-κB and upregulating the expression of microRNA-21 [127-129]. F. nucleatum induces apoptosis in lymphocytes, an ability mediated by heat-labile outer membrane protein(s), thus allowing the microorganisms to evade the immune system [130] (Fig. 3).
CONCLUSION
Chronic periodontal disease facilitates bacteria to alter the microenvironment in OSCC. The resulting inflammatory process modifies the immune response, inhibits cell apoptosis, stimulates angiogenesis, increases the IL-6 or TNF-α effects, and alters the cell cycle function. It has been found that bacteria such as P. gingivalis, or F. nucleatum differ quantitatively in each cancer stage, which could open an opportunity to consider these bacteria as possible diagnostic or prognostic markers in the management of OSCC. Moreover, oral native bacteria, such as Streptococcus and Neisseria could produce acetaldehyde affecting epithelial cell phenotype and its protective functions. This review also suggests the importance of raising awareness of prompt periodontal therapy and maintaining good oral hygiene by the dental care professional as a strategy for decreasing cancer risk. In summary, oral dysbiotic communities could promote oral cancer progression. However, cohort studies are still required to overcome statistical heterogeneity, bias, and confusion factors.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We want to thank Dr. Octavio Gonzalez from Kentucky University for his support and motivation with this research. And to Dr. Harvey Kessler (Retired) from Texas A&M School of Dentistry for his academic collaboration and loyal friendship.


