All published articles of this journal are available on ScienceDirect.
Bioinformatics and Data Mining Studies in Oral Genomics and Proteomics: New Trends and Challenges
Abstract
Genomics and proteomics have promised to change the practice of dentistry and oral pathology, allowing the identification and the characterization of risk factors and therapeutic targets at a molecular level. However, mass-scale molecular genomics and proteomics suffer from some pitfalls: gene/protein expression are significant only if inserted in a detailed network of molecular pathways and gene/gene, gene/protein and protein/protein interactions.
The proper analysis of these complex pictures requires the contribution of theoretical disciplines, like bioinformatics and data mining. In particular, data-mining of existing information could become a strong starting point to formulate new targeted hypotheses and to plan ad hoc experimentation.
In this review, advantages and disadvantages of the above-mentioned disciplines and their potential in oral pathology are discussed. The leader gene approach is a new data mining algorithm, recently applied to some oral diseases and their correlation with systemic conditions. The preliminary results of the application of the leader gene approach to the correlation between periodontitis and heart ischemia at a molecular level are presented for the first time.
“-OMICS” IN ORAL PATHOLOGY
Genomics have promised to change the practice of dentistry. Since the complete sequencing of the Human Genome, the development of mass-scale research tools in this field, such as DNA microarrays, has greatly improved [1]. This technique allows a complete profiling of the expression of a very high number of genes in a given tissue in a single experiment [2]. DNA microarrays can be used to measure the relative gene expression in different conditions, e.g. normal versus pathological tissues [2]. For instance, recent studies have employed microarray technology to monitor gene expression in periodontal patients [3-5]. In 2006, Steinberg et al. investigated the effects of interleukin-1beta on oral keratinocytes, using an epithelia-specific DNA microarray, and identified several genes associated to inflammatory response in periodontitis [3]. More recently, Vander-Sengul et al. reached similar conclusions, by analyzing more than 40,000 genes in a single experiment [4]. In another study, Beikler et al. have determined the expression profiles of several immune and inflammatory genes in periodontal tissues from patients affected by severe chronic periodontitis, either undergoing or not undergoing a periodontal therapy [5]. DNA microarrays have also been used to evaluate strain diversity of oral microflora implied in periodontitis [6,7] and the effect of oral microflora on the expression of chemokine genes [8].
These are only some examples of the potential of DNA microarray analysis in periodontitis. Similar studies have been conducted also in other diseases, such as oral lichen planus (OLP). A recent study by Tao et al. used DNA microarrays to characterize the gene expression profile of OLP lesions from nine patients, compared to nine healthy controls [9]. In total, 985 differentially expressed genes (629 up-regulated and 356 down-regulated) were identified, thus shading some new lights on the pathogenesis of OLP [9].
However, if compared to other areas of medicine, the progresses in oral pathology achieved using purely genomic approaches have been overall limited [10]. It has been suggested that other “-omics” disciplines, such as proteomics and metabolomics, should be applied to reach a deeper understanding of the molecular mechanisms underlying oral disorders [1].
In particular, proteomics deals with the identification of the proteins produced by cells in normal and diseased conditions, whereas metabolomics monitors the role of small molecules (lipids, sugars and amino acids) involved in daily cellular function [1]. The potential role of these disciplines in the study of oral biology and pathology appears immediate. For instance, proteomics of saliva can be used to evaluate the disease progression in periodontal patients [11]. Wu et al. have recently identified 11 proteins showing a different expression in the saliva of generalized aggressive periodontitis patients, when compared to healthy controls [11]. With respect to OLP, a metabolomic approach to the early disease diagnosis has been proposed [12]. In this study, saliva samples from patients and healthy donors were analyzed by High Pressure Liquid Chromatography (HPLC) and Mass Spectrometry (MS); then a diagnostic model was built using advanced bioinformatics and data-mining techniques [12]. The results have suggested that the metabolic profile may properly distinguish between oral squamous cell carcinoma (OSCC), OLP and oral leukoplakia from saliva samples. On these basis, a population-based screening of cancers and precancers lesions may be developed [12]. Noteworthy, this latter study shows also the importance of data-mining and bioinformatics in the interpretation of data emerging from genomics, proteomics and metabolomic studies.
THE INTEGRATION OF “-OMICS” DISCIPLINES IN ORAL PATHOLOGY
Many oral disorders, including periodontitis and OLP, are complex multifactorial, polygenic diseases [2]. In these diseases, the genetic etiology is not attributed to a single gene or to the encoded protein, but it is spread over several different genes, each having a modest effect. However, although each gene has a small effect, the overall effect of all the genes and of all the encoded proteins involved may be substantial [10]. Therefore, the pathophysiology of complex diseases is characterized by the involvement of various biologic pathways [2]. The identification and the characterization of candidate genes, proteins and molecules involved in a given disease by the integrated use of “-omics” disciplines will probably represent one of the milestones of future health care [13].
This integrated approach, also known as systems biology, allows to draw a picture of the expression levels of the candidate molecules associated to a given disease and of the interaction among them [13]. In fact, it has been suggested that a simple variation in expression of a single gene or of the encoded protein is not meaningful per se, but only if put in a proper framework of interactions (i.e. physical interaction of different molecules, involvement in the same metabolic patway, and co-expression in microarray studies) [14]. The analysis of the complex network of connections between genes/proteins may allow the identification of potential molecular markers and targets [13,14].
Gene interaction networks have recently been demonstrated and described in several biological processes [2,15-19]. In particular, genes showing the strongest interconnections with the other genes involved, i.e. hub or “leader” genes, may be supposed to play an important role in the specific process [14,15]. A small variation in the expression of these genes, which are often chromatin regulators, can deeply modify the effect of other genes involved in the process [13,17]. This approach will likely be extended from gene networks to gene/protein networks and, possibly, to other molecules playing a role in the process under study, further increasing the integration between the “-omics” disciplines.
The study of interaction networks requires the systematization and the analysis of a huge amount of information emerging from experimental studies. Theoretical disciplines, such as bioinformatics and data-mining, are therefore required to integrate this explosion of data [20,21].
BIOINFORMATICS AND DATA-MINING: INTEGRATING DATA IN A FRAMEWORK OF HYPOTHESES
There are approximately 30,000 genes in the human genome and the number of proteins is likely at least three times higher, as resulting from alternative splicing and post-translational modifications [20]. A complete experimental analysis of all the molecules involved in a given process, including both genes/proteins and small molecules, appears therefore an enormous task. Bioinformatics can become an added value in this context [17]. This discipline is defined as the application of information technology to the field of molecular biology, via the development of original algorithms. Originally developed for the analysis of gene sequences, bioinformatics is now widely applied to the study of gene and protein expression and to the analysis of genes and protein networks [21]. Years ago, it has been stated that “oral genomics will be transformed by the evolution of high-throughput techniques and "informatics"”, thus promising to change the practice of dentistry and oral pathology [21].
Although a similar revolution has not been observed yet, the importance of bioinformatics to analyze data and retrieve new information in oral “-omics” is beyond any doubt. However, like all disciplines, even bioinformatics suffers from some pitfalls. Let us consider, for instance, data analysis of microarrays: this process is usually performed with bioinformatics/statistical techniques, such as SAM (statistical analysis of microarrays) [17, 18]. However, many concerns have been raised against purely-statistical analysis of microarray, since microarrays are not a typical statistical object (few parameters, many samples) [18]. On the contrary, microarray investigation requires the correlation of a very high number of parameters with a very small number of samples (i.e. expression of many genes and few microarray slides analyzed) [18]. Therefore, some non-statistical methodologies of microarray analysis have now started to emerge [18]. It has been observed that purely-statistical analysis of microarray may not reveal low variations in gene expression [18]; however, even a small change in the expression of a given gene may result in a large effect downstream [17,18].
Moreover, the greatest part of genes displayed on an array is often not directly involved in the cellular process being studied [17]. Commercial arrays with a lower number of genes – usually 150–200 – are currently available, but the genes displayed are usually once again chosen without a precise consideration of the particular target of the study [17].
Another discipline playing an important role in the analysis of genomics/proteomics experiments is data mining, i.e. extracting patterns from data, thus developing new information from previous knowledge. Data mining commonly involves four classes of task (Table 1) [22]. With these methods, a further simplification of complex information emerging from genomics/proteomics experiments becomes possible. Properly combined with bioinformatics techniques and algorithms, data mining may allow to draw a simpler, but at the same time powerful picture of complex amount of data. However, data mining is only able to generate new well-grounded hypotheses, since it is completely based on previous information [2].
Different Classes of Task in Data Mining [22]
| Class | Task |
|---|---|
| Classification | Arrangement of a set data into predefined groups. Common algorithms include Nearest neighbor, Naive Bayes classifier and Neural network. |
| Clustering | Arrangement of a set data into not-predefined groups, grouping similar items together. Common algorithms include Hierarchical clustering and K-means clustering. |
| Regression | Definition of a function modelling data with the least error. A common method is Genetic Programming. |
| Association rule learning | Searching for relationships between variables. Common algorithms include Apriori algorithm and Eclat algorithm. |
Although there are some examples of the application of data mining techniques in dentistry [23,24], studies using data mining in oral pathology are still quite scant [2,14, 19,25].
DATA MINING STUDIES IN ORAL PATHOLOGY
In 2007, Hettne et al. applied different data mining techniques to generate and explore new hypotheses into how atherosclerosis and periodontitis could be linked at a genomic level, on the basis of previous literature and knowledge [25]. In total, 16 candidate genes were identified, with PON1 being the primary candidate. Therefore, further targeted studies may be planned specifically on candidate genes: such targeted analysis would be simpler than mass-scale molecular genomics, since it limits the number of objects, but at the same time powerful. Hettne et al. underlined also that collaborative efforts of investigators from different fields of expertise, such as oral pathology and data-mining, can result in the discovery of new hypotheses [25].
In the same period, and independently from the above-mentioned landmark study, the “Leader Gene approach” was first applied to oral pathology [14]. This data mining algorithm aims to identify all the genes involved or potentially involved in a given process and to rank them according to the number and confidence of interactions with the other genes, as derived from data mining of web-available databases [14]. Genes belonging to the highest rank are defined as “leader genes”, since they may be supposed to play a very important role in the analyzed processes. Therefore, as also suggested by Hettne et al., these genes can become the target of ad hoc experimentations. The leader gene approach gave promising results, confirmed experimentally, when applied to human T lymphocytes cell cycle [16,17], to osteogenesis [26], and to a complex model like kidney transplant tolerance [18].
In the field of oral pathology, the leader gene approach was first applied to periodontitis [2]. Via an interrelated and systematic query of several gene databases, 61 genes involved or potentially involved in periodontitis were identified. The interactions among these genes and among the encoded proteins were mapped and given a significance score using STRING, a web-available gene/protein interactions database [27]. Of note, only interactions with a high level of confidence (as provided by STRING database) [27] are considered in this analysis. The confidence level depends upon the experimental and statistical reliability of the particular study, or studies, indicating a given interaction. The calculation of a weighted number of links (weighted sum of scores for every interaction in which the given gene is involved) and subsequent clustering according to this parameter revealed that only 5 genes (NFKB1, RELA, PIK3R1, GRB2, CBL) had stronger interconnections with the other ones involved or potentially involved in periodontitis, as demonstrated by statistically comparing the weighted number of links. Of note, the involvement in periodontitis has been completely established for only two leader genes. Even with the limitations of any ab-initio analysis, this data mining study suggested a targeted experimentation on significant genes and, therefore, simpler than mass-scale molecular genomics. Such experimentation, conducted via Real-Time Polymerase Chain Reaction (RT-PCR) is now almost completed: the preliminary results indicate a significant, although limited, difference in expression of the leader genes in macrophages of periodontal patients and healthy controls (Marconcini 2009, personal communication). It must be emphasized that a mass-scale analysis of gene interactions would have failed in identifying such a limited variation in expression.
The leader gene approach can be applied to infer the correlation between oral diseases and between oral and systemic diseases at a genomic level [19]. For instance, a recent study evaluated the relationship between human periodontitis and type 2 diabetes: results showed that periodontitis and diabetes share four leader genes and that all leader genes are linked in a complex map of interactions. This finding may suggest an important role of leader genes in the association between these diseases; leader genes may be supposed, at least preliminarily, to act as hubs in the interaction map. As a further, although preliminarily confirm of these observations, a similar interconnection at a genomic level was not observed between periodontitis and a control disease, i.e. sinusitis, despite the aetiology of these conditions is similar [19]. At present, some similar studies, evaluating the correlation between OLP and OSCC, periodontitis and pemphigoid, and periodontitis and heart ischemia are ongoing; as an example of the method, data regarding the correlation between periodontitis and heart ischemia are preliminary presented here, for the first time.
CORRELATION BETWEEN HUMAN PERIODONTITIS AND HEART ISCHEMIA AT A GENOMIC LEVEL: A DATA-MINING STUDY
Periodontitis is a potential risk factor in patients with ischemic heart disease [28]. Among the several factors linking these two conditions, such as age, gender, presence of arterial hypertension or diabetes, smoking, diet, a genomic base has also been suggested [28]. The leader gene approach was applied to heart ischemia, according to the algorithm described in [2]; the resulting leader genes were compared, to formulate new hypotheses about the association between periodontitis and heart ischemia [19]. In total, 225 genes involved or potentially involved in human cardiac ischemia were identified. Among these genes, only 6 were invariably identified as leader genes, with an established or potential role in this disease (Table 2); their weighted number of links was significantly higher when compared to the other genes identified. Only 1 leader gene (NFKB1) is shared between periodontitis and heart ischemia (Table 2); however, leader genes of the two conditions are linked in a very close network (Fig. 1).
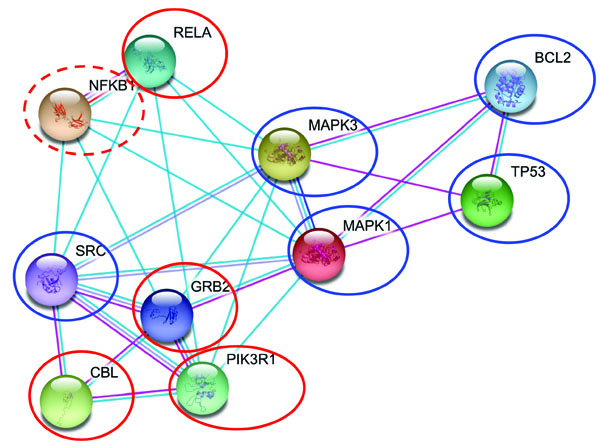
Interaction map of leader genes human heart ischemia (blue circles) and periodontitis (red circles). The lines which connect single genes represent physical interaction between proteins, confirmed by various experimental methods (magenta lines), correlation in gene expression experiments (dark blue) or the involvement in the same metabolic pathway (light blue). NFKB1, the shared leader gene is circled in dotted red.
Leader Genes and their Established or Putative Role in Periodontitis and Human Heart Ischemia. NFKB1, the Shared Leader Gene is Represented in Bold
| Periodontitis | Heart Ischemia | ||||
|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Role | Gene Symbol | Gene Name | Role |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | Increased activity beneath periodontal lesions | TP53 | tumor protein p53 | Induces apoptosis of cardiac myocytes |
| CBL | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Triggering of inflammation | NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
Some polymorphisms are associated to heart dysfunction
Triggering of inflammation |
| GRB2 | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Marker of severe periodontitis | BCL2 | B-cell CLL/lymphoma 2 | Protection of myocytes from apoptosis |
| PIK3R1 | growth factor receptor-bound protein 2 | Possible involvement in growth and differentiation in periodontal tissues, via the EGFR/RAS signalling | MAPK3 | mitogen-activated protein kinase 3 | Protection of myocytes from apoptosis |
| RELA | Cas-Br-M (murine) ecotropic retroviral transforming sequence | Possible involvement in bone resorption | MAPK1 | mitogen-activated protein kinase 1 | Protection of myocytes from apoptosis |
| SRC | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Induces apoptosis of cardiac myocytes | |||
How can these preliminary results be interpreted? NFKB1, the shared leader gene, is a established marker of inflammation in periodontitis [2] and some polymorphisms of this gene are associated to heart dysfunction [29]. Although these findings do require a confirmation and a deeper evaluation, this data mining study further suggests the importance of inflammation in the aetiology of both periodontitis and heart ischemia [28] and the importance of NFKB1 and of its neighbourhood in the interaction map in the association between the two conditions. These observations could lead to the planning of a targeted experimentation, focused on NFKB1 and on the other leader genes identified via the data-mining analysis. Such experimentation would be therefore focused on a limited number of genes likely having a great importance in the process. At the same time, the experimental complications and the difficulties in the purely-statistical analysis associated with mass-scale molecular genomics would be at least in part limited [2,14,17,18]. Possible means to conduct this targeted experimentation may be RT-PCR, thus allowing a high level of reliability, but studying a single gene at time, or targeted microarrays, with the possibility to infer co-expression of a small number of different genes. directly involved in the process being studied.
However, it should be stressed that the results of this data mining study must be considered more as well-supported hypotheses than as proven statements, since the entire process is based on previous knowledge. Data mining cannot provide new answers, but it can help us in formulating more targeted questions to be addressed experimentally.
THEORY PLUS EXPERIMENTS IN ORAL PATHOLOGY: TOWARDS THE FUTURE
Genomics and proteomics have promised to change the practice of dentistry and oral pathology; their potential to identify risk factors and therapeutic targets at a molecular level is established. However, mass-scale molecular genomics and proteomics suffer from some pitfalls: gene and protein expression is not significant per se, but only if inserted in a detailed cross-talk of molecular pathways and gene/gene, gene/protein and protein/protein interactions. The integration of genomics and proteomics, i.e. epigenetics, will likely provide many of the answers we still need in the understanding of the molecular basis of diseases.
However, the contribution of theoretical disciplines like bioinformatics and data mining will possibly become an added value in drawing such a complex picture [1]. In particular, data-mining of existing databases could be a starting point to formulate new hypotheses and to plan targeted experimentations, which may confirm or discard each hypothesis and suggest potential risk factors and therapy targets [19]. Current state-of-the-art in both “-omics” and theoretical disciplines suggests that a proper combination of experimental and theoretical results, obtained with different methods, will soon become the golden standard for the study of oral diseases.
ACKNOWLEDGMENTS
The authors declare no conflict of interest. The authors wish to thank Professor Francesco Chiappelli (UCLA, Los Angeles, USA), Dr. Victor Sivozhelezov (Nanoworld Institute, Genova, Italy) and Dr. Oluwadayo Oluwadara (UCLA, Los Angeles, USA) for useful discussion.


