All published articles of this journal are available on ScienceDirect.
In Vitro Evaluation of Enterococcus faecalis Adhesion on Various Endodontic Medicaments
Abstract
E. faecalis in endodontic infection represents a biofilm type of disease, which explains the bacteria’s resistance to various antimicrobial compounds and the subsequent failure after endodontic treatment. The purpose of this study was to compare antimicrobial activities and bacteria kinetic adhesion in vitro for three endodontic medicaments with a clinical isolate of E. faecalis. We devised a shake culture which contained the following intracanalar preparations: CPD, Endoidrox (EIX), PulpCanalSealer (PCS); these were immersed in a liquid culture medium inoculated with the microorganism. The shake system velocity was able to prevent non-specific bacteria adhesion and simulated the salivary flow. Specimens were collected daily (from both the medium and medicaments) for 10 days; the viable cells were counted by plate count, while the adhesion index AI° [E. faecalis fg DNA] /mm2 was evaluated in the pastes after DNA extraction, by quantitative real time PCR for the 16S rRNA gene. A partial growth inhibition, during the first 24 hours, was observed in the liquid medium and on the medicaments for EIX and subsequently for CPD (six logs). EIX showed the lowest adhesion coefficient (5*102 [fg DNA]/mm2) for nine days and was similar to the control. PCS showed no antimicrobial/antibiofilm properties. This showed that “calcium oxide” base compounds could be active against biofilm progression and at least in the short term (2-4 days) on E. faecalis cells growing in planktonic cultures.
INTRODUCTION
Enterococcus faecalis (Streptococcus faecalis) is a facultative anaerobic, Gram + cocci group D which is a member of the intestinal microflora [1]. It is currently isolated in a wide variety of human infections such as those of the urinary tract, the bloodstream, the abdomen, the endocardium and in situ foreign devices [2-5]. This organism is certainly the major cause of failure in endodontic treatments [6,7]. It may cause periapical disease, perhaps due to its ability to invade dentinal tubes remaining viable within the tubule; in addition, it is also able to adhere to collagen and may form a biofilm [7]. This organism is able to adhere to several surfaces with different kinetics in a different manner depending on the clinical isolate type [8,9]; the adherence mechanism and subsequent biofilm formation have an important role for bacterial colonization and survival in the host and are implicated in chronic infections [10]. In endodontic infection, E. faecalis first adheres to the tissue surfaces by a physical association; in a second step there is permanent bonding by specific bacterial adhesins to complementary receptors on the host surfaces. Once the bacterial cell is bound, it is able to use available nutrients and a biofilm structure is necessary to contend with host defense mechanisms and for resistance to antibacterial treatments. For these reasons, experimental data suggest that viable E. faecalis cells can be recovered from root canal systems after an effective chemo-mechanical instrumentation treatment [11-13]. However the coronal seal and the history of teeth harbouring enterococci have rarely been accurately investigated; a more likely recent explanation for the high occurrence of enteroccci in filled root canals is that they enter during or after treatment, the origin of this infection is most likely food [14]. In this contest E. faecalis intracanalar infection can be due to salivary contamination during treatment or to inadequate root canal obturation by salivary microleakages [15].
In this work we have studied the in vitro kinetic adhesion of E. faecalis in some endodontic treatments using traditional cultural methods and a novel integrated molecular procedure.
This technique consists of a particular dynamic shake culture system containing three different commercial intracanalar medicaments (see formula in material and methods): CPD EIX, PCS, immersed in a liquid medium and inoculated with the microorganism. The shake system velocity was able to oppose non-specific bacterial adhesion simulating the salivary flow in the oral cavity. The cell viability was evaluated by CFU enumeration, while the adhesion capacity (AI°) was evaluated by quantitative real time PCR; this molecular technology represents the most sensitive method for recognizing and quantitating bacterial DNA in a short time and with high precision. Here we introduce a method for E. faecalis adhesion measurement by the quantization of the 16S rRNA gene in these endodontic medicaments.
MATERIALS AND METHODOLOGY
Endontic Materials Used
Three different endodontic medicaments were used: (i) CPD, a recent medicament used in Italy, obtained by mixing three components: Stomilex (Stomigen-Rome, Italy) whose active principle is calcium oxide, Radiopaca iodoformic paste (Giovanni Ogna & Figli-Muggiò, Mi, Italy) whose active principles are iodoform, parachlorophenol, camphor, menthol and De Trey Zinc (Dentsply, De Trey-Konstanz, Germany); (ii) Endoidrox (EIX) whose active principle is calcium hydroxide; (iii) PulpCanalSealer, PCS (Kerr-USA, Endodontics), whose active principles are zinc oxide and Eugenol.
Each paste was made by following the manufacturer's recommendations and the three components of the CPD paste were mixed in equal parts. These compounds (using 0.25 mg of each Endodontic treatment) did not show any antibacterial activity with this clinical E. faecalis strain in 48 hours by using a conventional Kirby-Bauer diffusion method [16] (data not shown).
Strain Used in this Study
The Enterococcus faecalis strain used was a clinical isolate GO2, from endodontic specimens isolated from the Department of Dental Disease Prevention (University of Cagliari). The bacteria were cultured in Müller Hinton agar (MH Microbiol, UTA, Cagliari Italy), and identified by Api 20 Strep (bioMerieuxus Marcy l’etoile, France) [17]. A bacterial suspension was performed to obtain a concentration with a turbidity equivalent to the no. 3 McFarland standard (about 108 CFU/ml). Aliquots were stored at −70°C in Müller Hinton broth with 15% glycerol; this bacterial suspension was used to obtain a genomic DNA standard for a real time quantization curve, and to inoculate shake cultures.
Chemostat Shake Cultures
The dynamic culture model used (Fig. 1) [18, 19] was an apparatus composed as follows:

Schematic representation of the bascule culture system used in this work. The medium level and bacterial title (mean 107 cells/ml) were both maintained by the flow rate regulator setting valves.
- shaking table (Continental Instruments, Italy)
- a thermostatic 37° C chamber [Vivacar diagnostic, Vivadent, Italy] containing sterile flasks, Falcon 3024 (Becton Dickinson, Meylan Cedèx, France) each with 25 ml of Müller Hinton Broth (three flasks for each endodontic compound).
- each flask contained 10 glass concave supports (ø 9 mm) containing the same endodontic treatment (0.25 g), the concentration of the endodontic medicament in the medium per flask corresponded to 10%.
- two flasks contained glass vials without any endodontic medicament (negative control samples).
E. faecalis was maintained in a stationary phase for 10 days with a title range mean of 107 CFU/ml by supplying the flask with an input of new nutrients and the daily removal of liquid medium, 1/3 of total volume (Fig. 1).
We followed the idea that salivary flow rate plays a role in bacterial attachment in teeth and paste surfaces (i.e. during intracanalar restoration) [15]. For this reason the apparatus constructed for this experiment was able to generate a medium flow of approximately 0.5 ml/min on the medicament surface [20] following the subsequent formula:
[Ø = Sp/Sf*Mvf]
Ø = medium flow, ml/min in each paste surface
Sp = paste surface cm2
Sf = flask surface cm2
Mvf = medium flask volume moved in a min, ml/min
Method for Viable Cell Count
Viable bacteria were counted from the chemostat liquid medium and from each medicament by diluting samples and plating the dilutions (from 10-1 to 10-6) on Müller Hinton agar.
Each endodontic medicament surface was washed three times with 5 ml of sterile 0.9% NaCl solution and successfully scraped by curettage following the procedure indicated by Teles et al. for subgingival plaque [21]. The medicament was suspended in Müller Hinton Broth, vortexed and used for CFU enumeration as described before. The viable cells were expressed as colony-forming units per ml, CFU/ml.
DNA Extraction
Once a day for 10 days we collected three vials for each endodontic treatment. After four washes with nuclease-free H2O, the bacteria were removed from the surface by scraping on the medicament surface, as described previously; they were suspended in 400 µl of nuclease-free H2O to form a paste suspension. Genomic DNA was obtained by the CTAB modified method [22], using 400 µl of each sample (bacteria or paste suspension, After vigorous vortexing, 70 µl of sodium dodecyl sulfate (SDS) 10% and 5 µl of proteinase K 10 mg/ml (SIGMA–Aldrich, ST. Louis Missouri USA) were added; the mixture was incubated for 10 min. at 65°C. 100 µl of NaCl 5M and 100 µl of CTAB/NaCl (0, 274 M CTAB, and 0, 877 M NaCl) were subsequently added, and the mixture was then briefly vortexed and incubated at 65°C for 10 minutes. 750 µl of SEVAG (Chloroform:Isoamyl Alcohol 24:1, SIGMA) were added and the mixture was vortexed for 10 sec. After centifugation for 5 min (at 12000 rpm) 0.6 volumes of isopropanol (SIGMA) were added to the supernatant. After 30 min at –20°C and centrifugation for 30 min at 12.000 rpm, the pellet was dried at room temperature for 20 min and suspended in 15 µl of molecular biology grade distilled water (Gibco, Invitrogen, Paisley, Scotland, UK). E. faecalis genomic DNA obtained from the bacteria suspension was quantified by a RNA-DNA calculator (Pharmacia) and expressed as femtograms of E. faecalis DNA. 2 µl of this DNA suspension were used in the PCR real time reaction.
Real Time PCR
Real time PCR was performed using a LightCycler instrument and a LightCycler DNA Master SYBR Green I kit (Roche Diagnostics Mannheim Germany), according to the manufacturer’s instructions [23]. The 20 µl final volume contained: 4 mM MgCl2, 1 µM of each primer (OG33-OG123) and 2 µl of DNA extract. The PCR program was the following: (i) denaturation at 95°C for 30 sec, (ii) 40 cycles of 0 sec at 95 °C, 10 sec at 50 °C, 12 sec at 72°C, (iii) melting curve performed for 0 seconds at 95°C, 45°C, 95°C. Transition rates were 5°C/s in the 72°C segment, 0.1 °C/s in the 45°C segment and 20°C/s for another step. Fluorescence was detected at the end of the 72°C segment in the PCR step (single mode), and at the 45°C segment in the melting step (continuous mode) in the F1 channel. During initial optimization of real time reaction, PCR products were analyzed using agarose gel, to ensure correct sample product size, and by a melting curve analysis. The positive reaction showed a 90°C Tm peak.
Primers for Real Time PCR
Primers for real time PCR were designed using the 16S rRNA gene sequence, extracted from the NCBI database GenBank with accession number AY692453. Possible oligonucleotide dimer formation, self-complementarity and annealing temperatures of the primers were calculated using the Oligo program vers. 4 (MedProbe, Oslo, Norway). Real time PCR primers OG 33 (5’ –GACTACCAGGGTATCTAATC -3’) and OG 123 (5’- AGCAGCCGCGGTAATA -3’) amplified a region of 286 bp. The theoretical melting temperatures of these oligos (Tms) were calculated using module 1 of the DNA hybridization prediction algorithm program HYTHER http://ozone3.chem.wayne.edu/ [24] with the following sets of parameters: (i) Monovalent cation concentration at 0.05 mol/L, (ii) Mg2+ at 0.004 mol/L, (iii) a concentration of PCR products (Top/Bottom strands) at 10-7 mol/L and (iv) Hybridisation temperature at 37°C.
Expression of the E. faecalis DNA Concentration
A real time PCR standard curve was made on DNA extracts, obtained from different E. faecalis DNA suspensions with concentration ranges 107 -102 DNA fg/2 µl. The amount of bacterial DNA in the endodontic extracts was calculated by an interpolated threshold cycle with a standard curve (Y = -5,4X + 40,4, R2 = 0,98) [23]. We used the following equation to calculate the E. faecalis adhesion index:
AI°= ([DNA]*V°/2)/S°
AI° is the bacterial adhesion coefficient measured in fg DNA /mm2 endodontic compound surface.
[DNA]= fg DNA in 2 µl, calculated by PCR real time standard curve interpolation.
V°= volume of DNA extract suspension (15 µl during our experiments). S°= medicament surface (19.6 mm2).
RESULTS AND DISCUSSION
Recent hypotheses indicate that E. faecalis is introduced into the oral cavity in food or by operators during endodontic therapy. It can be spread by saliva in filled root canals during intracanalar treatments or by teeth lesions [14]; an association between biofilm formation and virulence has been reported for E. faecalis during dental root infection [25]. Its adherence to a dental surface is controlled by a number of variables including surface structures, nutrient availability, quorum sensing molecules and general fluid hydrodynamics [26]. Primary attachment of bacteria to a surface is mediated by non-specific interactions such as hydrophobic or Van der Wall’s forces, however with dental surfaces molecular adhesions are necessary. The liquid flow normally opposes non specific interactions but “at the same time” can be a stimulus for the transition from the planktonic, free-swimming, mode of existence to a mature biofilm [27,28]. We have made an in vitro model consisting of a shake E. faecalis culture containing three different endodontic medicaments. This system mimics an E. faecalis biofilm on endodontic sealers, and the medium flow/turbulence (simulating saliva in the oral cavity) was an important stimulus for biofilm modulation. In these experiments we observed two major events in nine days: (i) variations in microbial growth, (ii) variations in mass biofilm formation on the medicament surfaces. The antimicrobial activity of endodontic medicaments has been tested using various cultural methods [29]; these studies were however restricted to testing of the initial activity of the materials and their components and were tested by the diffusion Kirby Bauer method at the inhibitory concentrations. The concentrations used in medicaments reported in this work following the manufacturer’s instructions, showed no inhibitory effects on Müller Hinton in 48 hours with this clinical E. faecalis strain. However, as reported in Fig. (2), a good partial inhibition (six logs) within the first day, was observed on the MH liquid medium for CPD and EIX, with about 500 CFU/ml on 2*107 CFU in the medium without medicaments (control). CPS showed a 1 log max of inhibition at the first day culture and comparable results were observed by viable E. faecalis cell count on medicament surfaces; EIX showed the lowest CFU from the first to the fourth day (about 500 CFU/medicament). Subsequently the strain showed a progressive adaptation to the pastes (from the fourth day on). As has also been suggested by different authors [30] this microorganism showed a contradictory resistance profile against many endodontic medicaments, in particular with strains recovered in different growth phases or when specimens were compared; this explains the notable differences between different authors on comparing the same substance using different E. faecalis strains [31,32]. In our model E. faecalis showed the lowest adaptability or adhesion propriety with EIX followed by CPD; in the present paper these results have also been confirmed with molecular tools. Fig. (3) shows the E. faecalis DNA expressed as the adhesion index (AI) calculated by real time PCR with the three different compounds for nine culture days. This strain showed a different ability for biofilm formation and no bacterial DNA was detectable during the first hour of incubation (< 100 DNA copies/PCR); detection was however possible after 24 hours. PCS showed an increased bacterial adhesion during the first 48 hours with a subsequent decrease in the following days reaching its lowest point on the seventh day, after which there was a marked increase until the ninth day. The maximum adhesion index was reported on the ninth day: AI°=2.9*104. The adhesion of bacteria to the CPD paste remained at a low level for the first 48 hours, the highest count of total adhering bacteria was recorded on the seventh day: AI°=2.7*103 fg of E. faecalis DNA/mm2, (Fig. 3). EIX showed the lowest adhesion process, with the range and title between 200 and 500 fg E. faecalis DNA/mm2 over nine days. In this model, cultural (viable cells) and molecular (AI°) methods demonstrated that medicaments containing Calcium base compounds Ca(OH)2/CaO were most effective against microbial growth and against biofilm formation. This is in accordance with Baik et al. 2008 [33] who showed that calcium hydroxide activated E. faecalis lipoteichoic acid (LTA), considered a major virulence factor of gram-positive bacteria. Moreover, other authors have demonstrated that this substance in E. faecalis plays a positive role in biofilm formation due to its alanine esters [34].
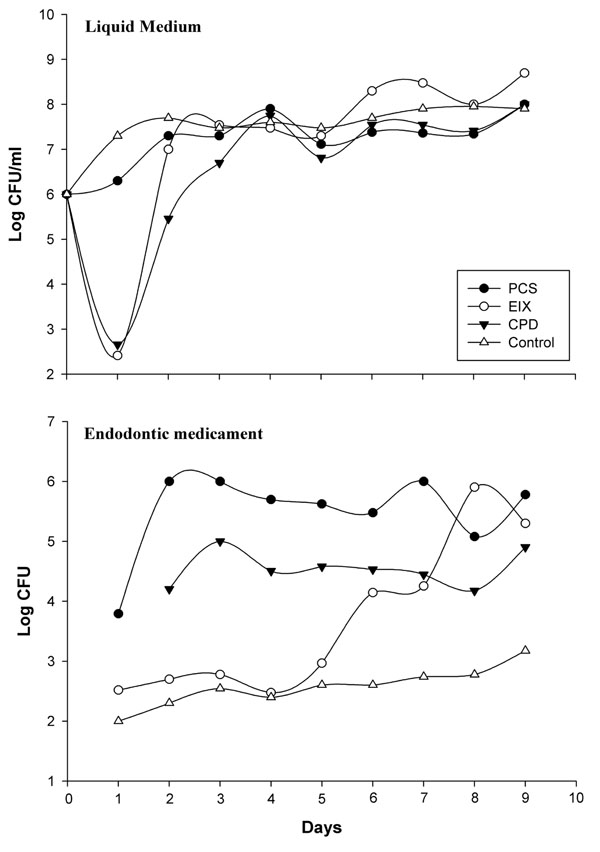
Growth curves of E. faecalis in medium and in each endodontic medicament reported for shake culture over nine day; each point represents the arithmetic mean of three determinations. The viable cells were expressed as colony-forming units in ml per liquid medium and in CFU/entire medicament surface (19.6 mm2).
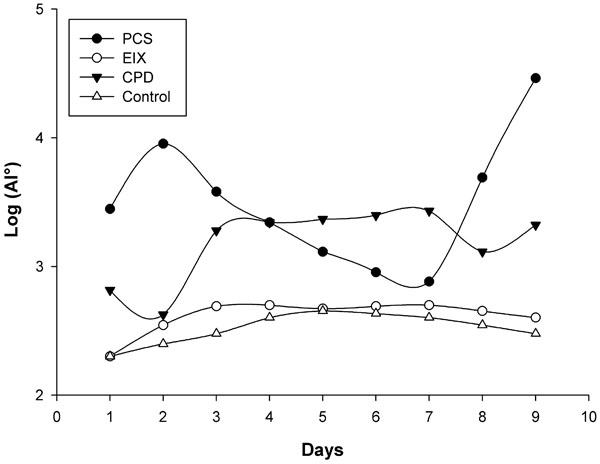
E. faecalis adhesion kinetic curves for the three endodontic compounds tested in this work. Means and S.D. of at least four experiments are shown.The control samples were concave supports without endodontic medicament and only the highest values are represented.
CONCLUSIONS
E. faecalis endodontic infection represents a biofilm type of disease where progression of the infection and treatment failure are due to the high adaptability of this bacterial biofilm towards endodontic active compounds. This study, using cultural and molecular techniques, supported the theory of E. faecalis’ ability to survive in these compounds; furthermore, it showed that calcium oxide base compounds could be active against biofilm progression, at least in the short term (2-4 days). This approach could be applied to new antimicrobials, as well as to "in vivo" analyses of teeth extracted after failed pulp therapy.


