All published articles of this journal are available on ScienceDirect.
L-lysine as a Potential Agent for Controlling Biofilm Formation Using Fusobacterium nucleatum and Porphyromonas gingivalis
Abstract
Background
Periodontitis, a globally-pervasive chronic inflammatory condition, is precipitated by forming microbial biofilms. A key player in this process is Fusobacterium nucleatum (F. nucleatum), an anaerobic bacterium exhibiting invasive and pro-inflammatory characteristics primarily associated with the onset of periodontitis. Consequently, developing novel biofilm control strategies specifically targeting F. nucleatum is essential.
The objective of this research is to examine the suppressive impact of L-lysine on biofilm formation using F. nucleatum.
Methods
Fusobacterium nucleatum(F. nucleatum) and Porphyromonas gingivalis (P. gingivalis) were cultured in a liquid medium containing L-lysine of different concentrations. The antimicrobial activity of L-lysine was evaluated using the broth microdilution method. The effect of L-lysine on the mono-species and dual-species F. nucleatum biofilms was assessed using techniques such as crystal violet staining. The synthesis of extracellular polysaccharide substances (EPSs) from biofilms was measured using phenol-sulfuric acid. The expression level of virulence factors (FN1420, FN0669, FN0634, FN1162) and the biofilm-formation-related gene radD were quantified using real-time PCR.
Results
L-lysine exhibited the antibacterial and antibiofilm activity of F. nucleatum and F. nucleatum/ P. gingivalis double strains, which was concentration-dependent. The MIC and MBC values were 10 mg/ml and 20 mg/ml in F. nucleatum single strains, while those in F. nucleatum/ P. gingivalis double strains were 20 mg/ml and 40 mg/ml. L-lysine with a concentration ≥20 mg/ml could reduce the EPS production in F. nucleatum and F. nucleatum/ P. gingivalis dual-species biofilms. L-lysine also inhibited the expression of genes encoding the F. nucleatum virulence factor (FN1420, FN0669, FN0634, FN1162) and the adhesive factor radD.
Conclusion
These findings corroborate the potential of L-lysine as an agent impeding the development of both mono-species biofilms of F. nucleatum and dual-species biofilms of F. nucleatum and P. gingivalis.
1. INTRODUCTION
Periodontitis is a chronic inflammatory disorder that can affect the supportive tissues of the periodontium, including the gums, periodontal ligaments, cementum of the roots, and the alveolar bone [1]. If left untreated, it will become the primary reason for tooth loss among adults [2, 3]. Although genetic, environmental and behavioral factors may contribute to the development of periodontitis, it has been scientifically confirmed that dental plaque biofilms play a significant role in the onset and occurrence of this condition.
Unlike planktonic bacteria, those living within biofilms demonstrate a heightened resistance to antibiotic treatments. The biofilms in dental plaque encase these bacteria and tolerate harsh conditions, resistance to antibacterial therapies, and evasion of the host's immune response [4]. Therefore, such dental plaque biofilms are a crucial focus for future research. Decades of research have identified many documented or putative virulence factors of P. gingivalis that may contribute to its persistence in the subgingival region. However, only now have we begun to understand how this Gram-negative asaccharolytic bacterium integrates its virulence attributes to enhance the pathogenicity of a polymicrobial community. It is now established that P. gingivalis-induced periodontitis requires the presence of the commensal microbiota, as P. gingivalis cannot cause periodontitis in germ-free mice despite colonizing this host [5]. The idea that P. gingivalis works together with other periodontal organisms is supported by animal models of periodontitis. These models have shown that when P. gingivalis is introduced alongside accessory pathogens or other keystone-like pathogens such as S. gordonii or T. forsythia, there is greater alveolar bone loss than when P. gingivalis is introduced alone. This synergistic effect may go beyond just manipulating the host's response and may involve cooperative communication between different species, which helps reinforce bacterial fitness and ultimately contributes to dysbiosis. Therefore, identifying synergistic bacteria for P. gingivalis is crucial in treating periodontal disease.
In 1998, Socransky et al. studied 185 subjects aged 51±16 years. This consisted of 160 individuals with periodontitis and 25 periodontally healthy individuals [6]. Gingival subgingival plaque samples were analyzed using whole-genomic DNA probes and DNA-DNA checkerboard hybridization techniques. The study compared and analyzed the categories and levels of common microorganisms in subgingival plaque samples from different groups with varying periodontal status. Based on these microbial groups' colonization and distribution patterns and their relationship with periodontal conditions, the researchers proposed the concept of a “periodontal microbial complex.” The second, “orange complex,” includes core bacterial groups closely associated with periodontitis, such as Tannerella forsythia and Fusobacterium nucleatum. It was found that Candida albicans, an opportunistic pathogenic yeast commonly found in the gastrointestinal tract and oral cavity, was able to coaggregate with F. nucleatum, facilitating its colonization [7]. Amer et al. reported that the colonization of Candida albicans in oral leukoplakia was correlated with an increase in F. nucleatum levels. This bacterium may indirectly increase the risk of oral cancer by exposing the oral mucosa to acetaldehyde produced by Candida albicans [8]. It was also discovered that Tannerella forsythia is often detected in subgingival plaque at sites of attachment loss in severe periodontitis, with a significantly higher detection rate in smokers. This bacterium can produce various toxic products and enzymes, such as protease-like enzymes, that can damage tissue. However, it is essential to note that these bacterial species do not act alone. They interact through surface molecules or secrete intercellular adhesion matrices, enabling them to grow and reproduce as a community. For example, although P. gingivalis is an acid-sensitive bacterium, F. nucleatum can create a more neutral environment by fermenting glutamate and asparagine, which favors the colonization of P. gingivalis [9]. Similarly, the colonization sites of Tannerella forsythia often contain F. nucleatum, and it is speculated that this relationship may be due to the F. nucleatum providing essential nutrients for Tannerella forsythia to grow.
Fusobacterium nucleatum (F. nucleatum) is one of the most commonly encountered species in biofilm formations, as it serves as a “bridge organism” between early and late colonizing bacteria in biofilms [10]. F. nucleatum produces multiple adhesins, including Fap2, RadD, and Aid1, which have been identified as playing a crucial role in facilitating interactions and coaggregation between F. nucleatum and a diverse range of microbial species (Streptococcus sanguinis, Tannerella forsythia, and Porphyromonas gingivalis) in the oral cavity. These adhesins are vital in forming dental plaque biofilm [11, 12]. As a result, new strategies for controlling subgingival dental plaque biofilms, specifically targeting F. nucleatum, are currently being explored in the most recent relevant trials. This is due to the need for improved management of plaque and the growth of F. nucleatum.
The investigation of exogenous amino acids as potential therapeutic agents for treating biofilms has received significant attention. Certain amino acids have been shown to affect both the metabolic processes and growth of biofilms [13-16]. ε-Poly-L-lysine is a natural antimicrobial cationic polypeptide consisting of 23-25 homogenous L-lysine. It is known to have antimicrobial effects against both Gram-positive and Gram-negative bacterial species [17]. Antimicrobial peptides (AMPs) are effective antibacterial agents with antiviral, antifungal, and antitumor activities [18]. In many studies, increased lysine/ amine composition enhances the relative composition of arginine residues or guanidinium moieties [19]. Our previous study demonstrated a decrease in the expression of a lysine-synthesis-related gene (FN1461) and an increase in the expression of genes (FN1868 and FN1869) related to lysine degradation in the biofilm of F. nucleatum compared to the planktonic condition. A preliminary in vitro study showed that L-lysine can exhibit antibacterial and antibiofilm activity against F. nucleatum. Reports have indicated that lysine itself could act as a potential agent to inhibit biofilm formation in teeth [20]. However, the above has not yet been explained and fully studied in periodontal area. The current research aims to evaluate the impact of L-lysine on the formation of biofilms using periodontal bacteria associated with gum disease. Two representative periodontal bacteria, P. gingivalis and F. nucleatum, were utilized for single-species and mixed-species biofilm experiments.
2. MATERIALS AND METHODS
2.1. Bacterial Strains, Media, and Growth Conditions
This study chose F. nucleatum (ATCC 25586) and P. gingivalis (ATCC 33277) as the standard reference strains. They were cultured on blood agar plates (Blood Agar Oxoid No. 2; Oxoid, Basingstoke, UK) enriched with 5% (v/v) pure sheep blood (Oxoid), 5.0 mg/L hemin (Sigma, St. Louis, MO, USA) and 1 mg/L vitamin K (Hopebio, Qingdao, China). The cultivation was conducted under anaerobic conditions (10% H2, 10% CO2 and balanced N2) at 37 °C for 3-5 days. Single colonies were selected and grown in a medium rich in proteins, specifically heart-brain infusion (BHI) (Hopebio, Qingdao, China), which was enhanced with 5.0 mg/L hemin and 1 mg/L vitamin K (Hopebio, Qingdao, China). This process continued until the colonies reached an exponential growth phase.
2.2. The Antibacterial Activity of L-lysine on F. nucleatum, F. nucleatum and P. gingivalis in a Planktonic State
The growth curves at 600 nm were determined using a SpectraMax M3 microplate reader 24 hours after inoculation with F. nucleatum at a concentration of 108 CFU/mL. The experiment was conducted on 96-well plates with various concentrations of L-lysine (0 mg/mL, 2.5 mg/mL, 5 mg/mL, 10 mg/mL, 20 mg/mL, 40 mg/mL, 80 mg/mL).
To assess the effectiveness of L-lysine against F. nucleatum and P. gingivalis in a liquid environment, 96-well plates were utilized. A co-incubation of F. nucleatum at 106 CFU/mL and P. gingivalis at 108 CFU/mL (100 μL/well) was used. The plates were supplemented with varying concentrations of L-lysine (0 mg/mL, 2.5 mg/mL, 5 mg/mL, 10 mg/mL, 20 mg/mL, 40 mg/mL, 80 mg/mL) and the absorbance at 600 nm was measured every 8 hours for a total of 72 hours.
2.3. The Antibiofilm Activity of L-Lysine in Mono-species F. nucleatum Biofilms as well as Dual-species F. nucleatum and P. gingivalis Biofilms
The crystal violet staining method was used to analyze the effects of L-lysine on F. nucleatum biofilms. F. nucleatum was grown in 24-well plates with varying concentrations of L-lysine (0 mg/ml, 2.5 mg/ml, 5 mg/ml, 10 mg/ml, 20 mg/ml, 40 mg/ml, and 80 mg/ml) for 24, 36, and 48 hours. These plates were used as a control for the assay. Each well was rinsed thrice with PBS to remove unattached bacteria and then air-dried for 20 minutes. The wells were then treated with 2.5% glutaraldehyde for 15 minutes and stained with 0.1% crystal violet for 15 minutes at room temperature. The unattached dye was washed away with PBS, and the remaining solution was decolorized with 95% ethanol. The solution was then transferred to a 96-well plate, and the absorbance was measured using a microplate reader at a wavelength of 490 nm.
To evaluate the antibiofilm properties of L-lysine against dual-species F. nucleatum and P. gingivalis biofilms, F. nucleatum was co-cultured with P. gingivalis in 24-well plates at a density of 106 CFU/mL and 108 CFU/mL, respectively. Different concentrations of L-lysine (0 mg/ml, 2.5 mg/ml, 5 mg/ml, 10 mg/ml, 20 mg/ml, 40 mg/ml, and 80 mg/ml) were added and the crystal violet staining protocols described above were followed.
2.4. Measurements of Extracellular Polysaccharide (EPS) Using the Phenol-sulfuric Acid Method
We used the phenol-sulfuric acid method to evaluate the effect of L-lysine on the extracellular polysaccharide (EPS) of single-species F. nucleatum biofilms. Both single and dual-species F. nucleatum and P. gingivalis biofilms were grown using the previously described conditions. Afterward, the F. nucleatum biofilms were washed three times with PBS. Next, a solution of 10% phenol (40μL, diluted in distilled water) and 97% sulfuric acid was added, followed by a 10-minute drying period at room temperature. Finally, the EPS was measured by recording the absorbance at an OD of 490 nm.
2.5. Analysis of Biofilm Structure Using Confocal Laser Scanning Microscopy (CLSM) and Scanning Electron Microscopy (SEM)
The effects of L-lysine on single-species F. nucleatum biofilms and dual-species F. nucleatum and P. gingivalis biofilms were evaluated using laser scanning confocal microscopy (CLSM) and scanning electron microscopy (SEM). F. nucleatum was plated at a concentration of 108 CFU/mL in 24-well plates supplemented with varying concentrations of L-lysine (0 mg/ml, 2.5 mg/ml, 5 mg/ml, 10 mg/ml, 20 mg/ml, 40 mg/ml, 80 mg/ml) for 24, 36, and 48 hours.
Then, a dye solution containing SYTO9 and PI at a ratio of 1:1 was left in the dark for 15 minutes before being removed. The sample was then rinsed three times with sterile saline and placed upside down in a confocal viewing cuvette for evaluation using a laser scanning confocal microscope. Bacteria were visualized using a fluorescence microscope (Nikon ECLIPSE, TS100) and a laser-scanning confocal microscope (CLSM; A1R, Nikon, Japan). To observe the dual-species biofilms, F. nucleatum at a density of 106 CFU/mL was co-cultivated with P. gingivalis at a density of 108 CFU/mL in 24-well plates supplemented with varying concentrations of L-lysine for 24, 36, and 48 hours. The dual-species biofilms were then analyzed using the CLSM above.
Moreover, SEM was used to observe both single-species F. nucleatum biofilms and dual-species F. nucleatum and P. gingivalis biofilms, all cultured for 24 hours with varying concentrations of L-lysine. The samples were then fixed in 4% paraformaldehyde at 4 °C for 4 hours, washed three times with PBS, and dehydrated using increasingly concentrated ethanol solutions. After fully drying at the critical point, the samples were coated with a thin layer of gold. Subsequently, the structure and characteristics of these gold-coated samples were analyzed using a scanning electron microscope (JSM6400; JEOL, Tokyo, Japan).
2.6. RT-qPCR Analysis
We aimed to investigate how L-lysine influences the expression of genes that encode the virulence factor of F. nucleatum (specifically, FN1420, FN0669, FN0634, and FN1162) and the adhesion factor (also known as radD, or FN1526) [21]. F. nucleatum was seeded at a 108 CFU/mL concentration in 48-well plates and supplemented with L-lysine at varying concentrations for 6 hours. Total RNA was extracted using TRIzol® reagent (Ambion, Life Technologies, Carlsbad, CA, USA) following the manufacturer's guidelines and consistent with previous methods. The RNA was then converted into cDNA for sequencing using a HiScript III 1st Strand cDNA synthesis kit (R312-01, Vazyme, Nanjing, China). RT-qPCR reactions were performed using an iQTM SYBR® Green Supermix (Q511-02, Vazyme, Nanjing, China), with the specific primers shown in Table 1. The thermal cycling process included an initial step at 95 °C for 30 seconds, followed by 40 cycles at 95 °C for 10 seconds and 60 °C for 30 seconds, and ending with a sequence at 95 °C for 15 seconds, 60 °C for 1 minute, and 95 °C for 15 seconds. Relative transcription levels were calculated using 2-ΔΔt.
To determine the effect of L-lysine on the gene expression of the adhesion factor of F. nucleatum in dual-species biofilms of F. nucleatum and P. gingivalis, we co-incubated F. nucleatum (at a density of 106 CFU/mL) and P. gingivalis (at a density of 108 CFU/mL) in 48-well plates for 6 hours, while varying the concentration of L-lysine. RT-qPCR was performed using previously described methods, with the specific primers in Table 1.
2.7. Statistical Methods
A statistical evaluation was performed using GraphPad Prism 9. The normality of the data and the homogeneity of variance were confirmed through the Kolmogorov– Smirnov test and the modified Levene’s test, respectively. A one-way analysis of variance (ANOVA) was conducted to analyze the differences between multiple groups, and Tukey’s test was used for multiple comparisons. Any significant differences were determined with a significance level (α) 0.05. The microbial tests were repeated three times.
3. RESULT AND DISCUSSION
3.1. Statistical Analysis
3.1.1. L-lysine shows Antibacterial Activity against F. nucleatum, F. nucleatum and P. gingivalis
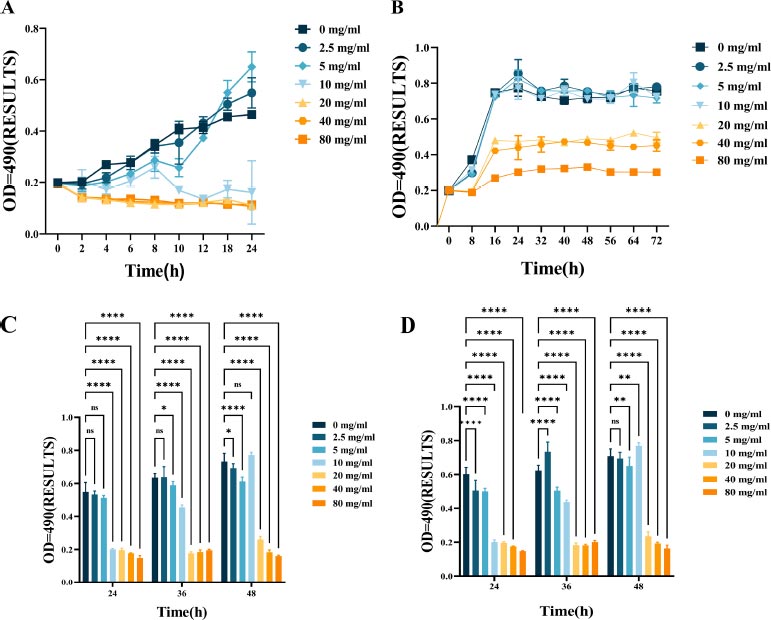
Fig. (1) shows the bacteriostatic effect of L-lysine treatment on F. nucleatum. This is evident from the growth patterns of F. nucleatum with varying concentrations of L-lysine over some time. The bacteria's growth was significantly inhibited when treated with 10 mg/ml of L-lysine compared to the control group (Fig. 1A). The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values of F. nucleatum with L-lysine were determined to be 10 mg/ml and 20 mg/ml, respectively. However, there was no noticeable change in the growth curve when the concentration of L-lysine was below 10 mg/ml.
| GENE | Reverse Sequence 5’-3’ | Forward Sequence 3’-5’ |
|---|---|---|
| 16s | ACTGTTAGCAACTACCGATGT | TGTAGATGACTGATGGTGAAA |
| FN1420 | GAAGGTTGTGGTCATCCAGAT | CAAACATTGCTCCACCCATTA |
| FN1162 | TGTGGAGGACGCAATTTAGAA | ACCGATAGTATCTCCAGGAGT |
| FN0669 | GAGTTTGATGGCACTTGTAGG | GTCCACCTGAAAGCTCTGATA |
| FN0634 | GGTTCCAGGTCAAGATGGTTA | CAGAACCAGCTGCTCTCATAT |
| RadD | GGATTTATCTTTGCTAATTGG | ACTATTCCATATTCTCCATAA |
The growth curves of F. nucleatum and P. gingivalis are shown in Fig. (1B), along with the growth inhibitory effect of L-lysine. It was also discovered that L-lysine, at a concentration of ≥20 mg/ml, inhibited the growth of both F. nucleatum and P. gingivalis. The MIC of F. nucleatum and P. gingivalis was recorded as 20 mg/ml when using L-lysine, and their MBC value was recorded as 40 mg/ml.
The effect of L-lysine on the formation of single- and dual-species F. nucleatum biofilms was assessed using CV staining. As the concentration of L-lysine was increased, there was a noticeable reduction in the formation of F. nucleatum biofilms. This reduction went from 0.22 ± 0.0023 at 0% to 0.08 ± 0.0085 at 8% after a 24-hour incubation period. Lysine was used as a control in this experiment (Fig. 1C). Furthermore, the results from CV staining showed that at a concentration of 40 mg/ml or higher, L-lysine effectively prevented the formation of both F. nucleatum biofilms and dual-species P. gingivalis biofilms (Fig. 1D).
3.2. EPS Degradation within Mono-species and Dual-species F. nucleatum Biofilms
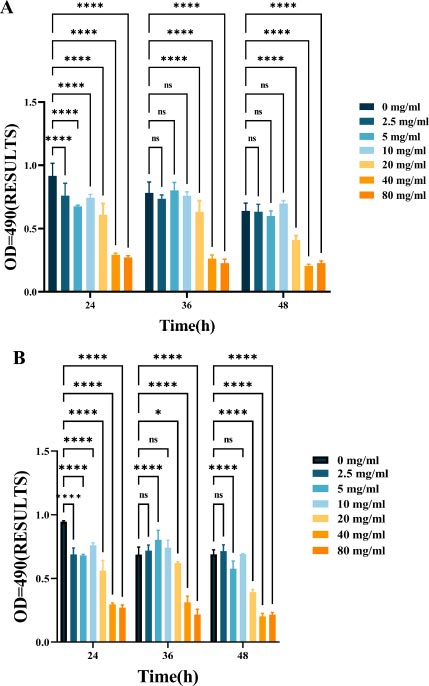
As shown in Fig. (2), L-lysine can effectively suppress EPS production in mono-species F. nucleatum biofilms. As the concentration of L-lysine increased, there was a corresponding decrease in EPS production, with levels dropping from 0.94 ± 0.099 to 0.27 ± 0.013 over a 24-hour incubation period (Fig. 2A). L-lysine also had a similar inhibitory effect on dual-species F. nucleatum biofilms, in addition to P. gingivalis, and a similar trend was observed after 36 and 48 hours. As the concentration of L-lysine increased, there was a significant decrease in the EPS content, dropping from 0.56 ± 0.043 to 0.17 ± 0.01 (Fig. 4B).
3.3. L-lysine Shows Antibiofilm Activity against Mono-species and Dual-species F. nucleatum Biofilms
This research aimed to examine the effect of L-lysine on forming F. nucleatum biofilms, both as a single-species and in combination with other species. It was found that L-lysine at a concentration of ≥20 mg/ml could inhibit the formation of F. nucleatum biofilms, as shown in Fig. (3A). L-lysine effectively prevented the formation of single-species F. nucleatum biofilms at different time points, with a similar trend seen after 36 and 48 hours. Without L-lysine, the biofilms formed by F. nucleatum were consistently distributed and fully covered the surface to which the bacteria were attached. However, when L-lysine was added, the biofilms were looser and tended to aggregate into smaller clusters (Fig. 3A). SEM analysis confirmed that F. nucleatum created thick, three-dimensional biofilms in a single-species environment. The bacteria in these biofilms displayed their typical spindle-like, rod-like, or fusiform shapes with varying lengths. Some bacteria were organized in multiple layers. The addition of L-lysine significantly reduced the number of F. nucleatum bacteria and damaged the structure of the biofilms, as seen in both the CLSM and SEM results (Fig. 4A).
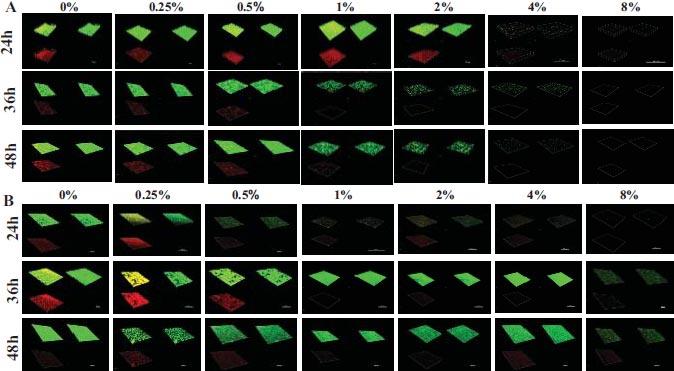
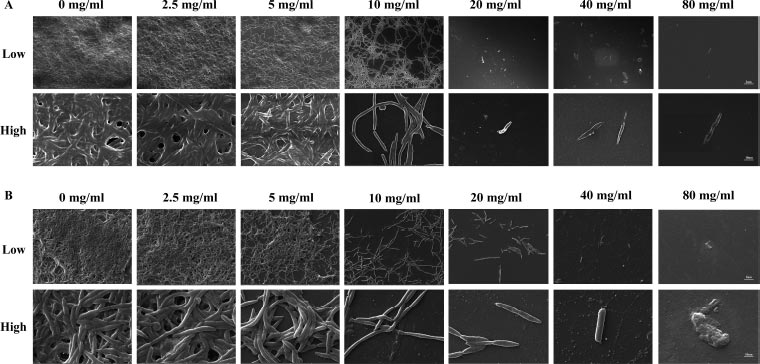
It also has been shown that L-lysine has a comparable inhibitory effect on dual-species biofilms formed by F. nucleatum and P. gingivalis. CLSM found that biofilms treated with L-lysine had a significantly reduced thickness (Fig. 3B). SEM analysis also showed that L-lysine could decrease the number of F. nucleatum and P. gingivalis bacteria present and alter the structure of the biofilms (Fig. 4B).
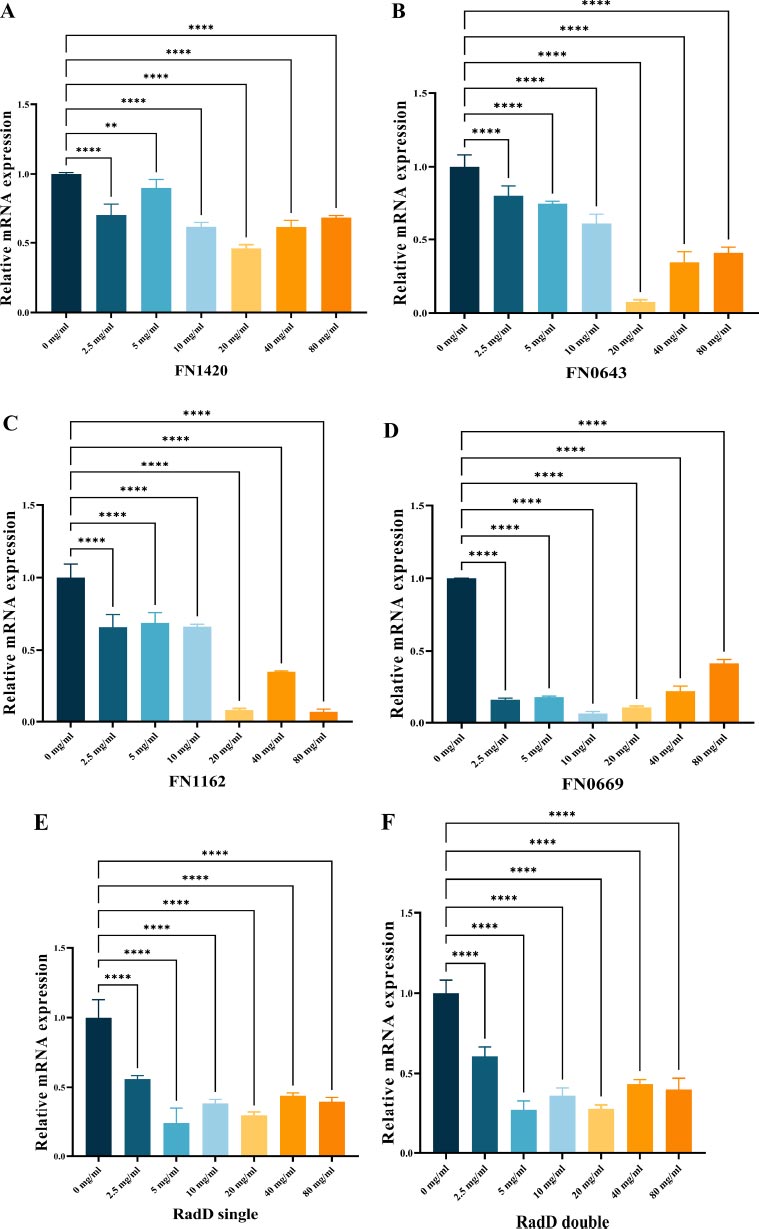
3.4. Gene Expression in the Biofilms
To investigate gene expression, we used RT-qPCR analysis as a quantitative method to measure the impact of L-lysine on biofilms produced by F. nucleatum. Our selection of genes included FN1420, which encodes the Na+/H+ antiporter NHAC; FN0669, which encodes the ATP binding protein znuC; FN0634, which encodes the GTP binding protein TypA/BipA; and FN1162, which encodes the enzyme hydroxyacylglutathione hydrolase. Our results showed that L-lysine effectively reduced the expression of all four genes studied in F. nucleatum biofilms. Interestingly, as the concentration of L-lysine exceeded 20 mg/ml, there was a corresponding decrease in the fold change of gene transcripts. At 20 mg/ml of L-lysine, a significant reduction in the expression of these four genes was observed (Fig. 5A-D).
As shown in Fig. (5E, F), radD encoding the RadD protein was significantly down-regulated in both single-species F. nucleatum biofilms and dual-species F. nucleatum and P. gingivalis biofilms in the presence of L-lysine.
4. DISCUSSION
F. nucleatum is a “bridging bacterium” commonly found in both supra-gingival and subgingival dental plaque biofilms. It has been shown to play a crucial role in their development [22, 23]. The eradication of F. nucleatum is a therapeutic strategy for periodontitis. This is predicted to lead to a decrease in subgingival biofilm mass and inflammation [24]. L-lysine was chosen because it possesses positively charged cationic amino acid groups that are highly effective in disrupting bacterial membranes, leading to the elimination of the bacteria [25, 26]. This study aimed to examine the effects of L-lysine on single- and dual-species F. nucleatum biofilms. Results showed that at a concentration of ≥20 mg/ml, L-lysine can hinder the formation of both single- and dual-species F. nucleatum biofilms. CLSM and SEM analysis showed a considerable decrease in bacterial count and biofilm thickness using L-lysine treatment. This treatment also led to a significant reduction in the amount of EPS present in the biofilm. Overall, these findings verify the potential use of L-lysine to control biofilm formation in F. nucleatum.
The microbial cell wall is the primary protective layer of the cell, providing both osmotic and physical protection [27]. Zhilei Tian et al. discovered that the main mechanism of action of ε-Poly-L-lysine on the cell wall of S. cerevisiae is to make the cell wall more fragile. This leads to an increase in cell wall permeability, allowing ε-Poly-L-lysine to enter the cells. As a result, the protoplasm leaks through the pores, ultimately causing cell death. Additionally, ε-PL can cause intracellular accumulation of ROS and DNA fragmentation in S. cerevisiae, contributing to cell death. These above findings might provide insight into the antimicrobial effect mechanism of L-lysine [28].
In previous studies, L-lysine has also been identified as a competitive inhibitor of F. nucleatum auto-aggregation and coaggregation. The inhibitory effect of L-lysine on Prevotella nigrescens and Capnocytophaga ochracea can be attributed to the ability of F. nucleatum to adhere to certain gram-negative oral bacteria, such as Prevotella intermedia [29-31]. Wu and his team performed thorough transposon mutagenesis of the fusobacterial genome to identify factors contributing to coaggregation. Their findings emphasized the interconnected roles of two components, the CarRS two-component signal transduction system and a lysine metabolic pathway controlling the crucial coaggregation factor radD. Transcriptome analysis revealed that CarR regulates a broad regulon, including metabolic genes such as radD and lysine, specifically kamA and kamD. The expression of these genes was significantly increased in mice lacking the carR gene, suggesting that lysine serves as a metabolic inhibitor of coaggregation and radD acts as a receptor [32]. The radD gene encodes the F. nucleatum radD adhesin, which plays a crucial role in cross-species adhesion and is integral to the structured architecture of multi-species biofilm [21]. This study aimed to investigate the cause of the effect of L-lysine on F. nucleatum biofilms, both single- and dual-species. RT-PCR assays showed that radD was noticeably inhibited in both types of biofilms after treatment with L-lysine. Our findings suggest that L-lysine may function as a metabolic inhibitor of coaggregation, with RadD acting as a receptor.
In parallel, we aimed to evaluate the effect of L-lysine on several virulence factors of F. nucleatum, including the FN1420, FN0669, FN0634 and FN1162 genes. We observed a decrease in the expression of these genes in F. nucleatum of a biofilm state with increasing levels of L-lysine treatment. Merritt et al. found that L-lysine was also capable of inhibiting the expression of FN1989 (encoding sodium-dependent tyrosine transporter), FN1988 (encoding tyrosine phenol-lyase), FN1586 (encoding O-succinylbenzoate-CoA synthase) and FN0941 (encoding gamma-glutamyltranspeptidase) with F. nucleatum in a planktonic state [25].
CONCLUSION
This study shows that L-lysine significantly prevents biofilm formation of both single- and dual-species F. nucleatum biofilms. As a result, L-lysine is being considered as a potential therapeutic option for controlling oral biofilms. Further research is needed to understand better the molecular processes contributing to biofilm disruption. One limitation of this study is the use of dual-species biofilms for analysis. However, the oral microbiome is highly complex, and considering additional bacterial species in future studies would provide a more comprehensive understanding. Therefore, future clinical studies on complex biofilms are necessary to demonstrate the effectiveness of L-lysine. Another limitation is that only one D-amino acid has been proven effective. Thus, further studies are needed to evaluate the combined effects of multiple D-amino acids for F. nucleatum or multi-species periodontal biofilms. In conclusion, our findings suggest combining various lysine forms may have a positive impact. While L-lysine shows promise as a biofilm control agent, addressing these considerations in future studies will be crucial for advancing its potential role in periodontitis management and improving oral health outcomes.
LIST OF ABBREVIATIONS
| BHI | = Heart-brain infusion |
| AMPs | = Antimicrobial peptides |
| EPS | = Extracellular Polysaccharide |
| CLSM | = Confocal Laser Scanning Microscopy |
| SEM | = Scanning Electron Microscopy |
| CLSM | = Confocal microscopy |
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
AVAILABILITY OF DATA AND MATERIALS
The raw/processed data required to reproduce these findings can be shared now.


