Bioinformatics Analysis of the Rothia dentocariosa Proteome and Assessment of the Proinflammatory Potential of Biofilm and Planktonic Cells
Abstract
Background:
Rothia dentocariosa is an opportunistic pathogen found in the oral cavity and is found to be involved in many oral infections as it has the ability to attach to the tooth and mucosal surfaces, produce substantial amounts of acids and integrate into dental plaque biofilms.
Aim:
To analyze the proteome of R. dentocariosa by using bioinformatics tools and to investigate the proinflammatory potential of R. dentocariosa.
Materials and Methods:
Protein sequences of R. dentocariosa were downloaded from NCBI and various in silico analyses were performed using bioinformatics tools. R. dentocariosa CCUG 35437 was grown on blood agar in 5%CO2 in air at 37 C for 2 days. Biofilms were cultured for 2 days and quantified by crystal violet staining. Human whole blood was stimulated with biofilms, biofilm-supernatants, planktonic cells, and whole cells. Proteome Profiler and ELISA-based quantification of cytokines were performed for the samples.
Results:
In silico analysis of the whole genome and proteome of R. dentocariosa revealed a number of proteins predicted to be potentially secreted but also possess virulence properties. R. dentocariosa was able to form only moderate biofilms. The ability of R. dentocariosa to induce different cytokines varied depending on the stimulant being used. Biofilms and planktonic cultures induced specific cytokines that were not induced by whole cells or biofilm supernatants. While IL-8 was induced at near-similar levels from biofilm and planktonic cells, IL-10 was induced at significantly higher levels (P<0.05) only by the planktonic cultures. The biofilm-supernatant and the whole cell stimulants induced lower levels of cytokines than biofilm and planktonic cultures.
Conclusion:
Identification of potential virulence factors predicted to be secreted extracellularly may suggest a key role for R. dentocariosa in oral and non-oral infections. Different stimulants from R. dentocariosa showed varying potential to induce cytokines from human whole blood. This may suggest differences in the composition/concentration of the bacterial components in the stimulants, with varying abilities to induce cytokine production, maybe the reason for the observed differences.
1. INTRODUCTION
Rothia dentocariosa is a pleomorphic gram-positive, aerobic nonmotile, non-spore-forming bacterium, which is commonly found as a member of the normal oral microbiota. It is an opportunistic pathogen implicated in oral infections such as caries, periodontitis, peri-implantitis [1, 2]. Further, the role of R. dentocariosa in causing endocarditis, pneumonia, and infections of peritoneum has also been reported [3-6].
Owing to its ability to attach to tooth surfaces, produce large amounts of acids, and get integrated into polymicrobial dental plaque biofilms, R. dentocariosa is regarded as a potent cariogenic species [2]. R. dentocariosa possesses a diverse set of virulence factors, whose role has not been fully elucidated. A few earlier studies have shown that R. dentocariosa stimulates TNF-alpha in a TLR-2-dependent mechanism [7]. Investigating the components of the complete inflammasome in the inflammation may shed more light on the pathogenesis mechanisms of R. dentocariosa.
The aim of this study was to perform in silico analysis of the proteome of R. dentocariosa using bioinformatics tools and to investigate the effect of R. dentocariosa biofilms-secreted components on cytokine production from human whole blood. Recent knowledge indicates that the virulence capabilities of bacterial species are exacerbated in their biofilm lifestyle [8]. Therefore, it is of significance to investigate the proinflammatory potential of different components of R. dentocariosa biofilms, biofilm-supernatants, planktonic cultures, and whole-cell preparations.
2. MATERIALS AND METHODS
2.1. Bioinformatics Analyses
The complete genome sequence and annotated protein sequences (in FASTA format) of Rothia dentocariosa ATCC 17931 were downloaded from NCBI databases. Signal-peptide bearing proteins were predicted by using the online tool SignalP, version 5.0 [9]. Lipoproteins in the secretomes were predicted using the prediction tools LipoP and PRED-LIPO [10]. Further, the prediction tool TatP was used to predict proteins secreted via the Twin-arginine translocation pathway (Tat-pathway) [11]. The subcellular localization of the proteins was predicted using the PSORTb tool, version 3.0.2 [12]
VirulentPred and MP3 (Predict Pathogenic Proteins; http://metagenomics.iiserb.ac.in/mp3/index.php was utilized to predict potent virulence factors in the secretomes [13], along with the Virulence Factor Data Base (VFDB). Gene ontology analyses were performed using Blast2Go and Cello2GO tools.
2.2. Reference Bacteria, Culture Conditions and Crystal Violet Staining
Reference strain R. dentocariosa CCUG 35437 purchased from Culture Collection University of Gothenburg was cultured on brucella blood agar in 5% CO2 in the air at 37oC for 2 days. After incubating for 2 days, the colony morphology on the agar plate was checked for purity under stereomicroscope and further by Gram staining.
2.3. Biofilm Culture and Biomass Quantification
For biofilm culture [14], the bacterial cells were first harvested from agar plates using a sterile swab and homogenous cell suspension was prepared in 1 ml of sterile PBS. The suspension was vortexed, and washed twice in sterile PBS by centrifugation at 5000 ×g for 5 min and the cell pellet was collected after the supernatant removal. For optical density (OD600) measurements, the homogenous bacterial suspensions from the stock suspension were 10-fold diluted in sterile PBS. OD600= 1 cell suspension was prepared as per Oral Microbiology Laboratory; Faculty of Dentistry; Kuwait University standard protocols and spectrophotometer was used to measure OD. Biofilm culture was initiated by inoculating 4 wells of the 24-well plates by adding 900 µl brucella broth and 100 µl aliquot from the OD600 = 1 bacterial cell suspension in each well. As a negative control, wells containing broth only but no bacteria were used in each experiment. All experiments were run in duplicate and three independent experiments were performed. For quantifying the biomass, the supernatant broth was removed, and the 2-day old biofilm was washed twice with 1 ml sterile PBS to remove the unattached cells. Methanol (1ml per well) was added and kept undisturbed in a fume hood at room temperature for 15 minutes. Methanol was then removed and the 24 well plate was kept open to dry at room temperature for 45 minutes. After that, 1 ml of 0.1% aqueous crystal violet stain solution was added to the wells and incubated for 20 minutes at room temperature. The stain was removed, and the 24-well plate was washed at least 7 times with tap water to ensure the complete removal of the stain. To remove the excess water from the wells, the plate was tapped on a dry tissue paper and then kept open in the fume hood for 5 minutes at room temperature, after which crystal violet-stained biomass photos were captured. Further, 500 µl of 33% acetic acid was added to each of the biomass-stained wells for destaining and kept on a shaker for 5 minutes. Three hundred microliter from each well of the 24-well plate was transferred into 3 of the wells of 96-well plate (triplicates), each containing 100 µl. Quantification was done by measuring the optical density (590 nm) using a spectrophotometer.
2.4. Stimulation of Human Whole Blood with R. dentocariosa Biofilm Cultures, Planktonic Cultures and Whole Cell Preparations:
2.4.1. Biofilm Cultures, Planktonic Cultures and Whole Cell Preparations of R. dentocariosa.
Biofilm cultures were set up in 24-well cell culture plates (BD Bioscience) as described in the previous section [14]. For planktonic and whole cell preparations (WCP), 100-µl aliquot from OD600 = 1 suspension was inoculated into 1.5 ml Eppendorf tubes containing 900 µl brucella broth. The 24-well plate and Eppendorf tubes containing cultures and control broths for biofilm and planktonic cultures were incubated for 24 h in 5% CO2 in air at 37°C for biofilm formation. As a negative control, wells and tubes containing broth only but no bacteria were used in each experiment. All the experiments were run in duplicate and two independent experiments were performed. Tubes containing WDC preparations were stored in -80o C freezer. After 24 hours of incubation, the supernatants of biofilm cultures in wells, planktonic broth cultures and whole cell preparations were removed, filter sterilized through a 0.2 µM syringe filter and then transferred to new sterile Eppendorf tubes. These filtered samples of R. dentocariosa were further used for stimulating human whole blood.
2.4.2. Stimulation of Human Whole Blood with Biofilms, Planktonic Cells and Whole Cell Preparations
The Kuwait University Health Sciences Center's Ethical Committee approved this study (DR/EC/3413). Human whole blood was collected from a systemically healthy human volunteer by venipuncture into (EDTA) tubes containing a heparin vacutainer (4 ml per tube). The whole blood was stimulated with live static biofilms, biofilm-supernatants, planktonic cells, or whole cell preparation for 24 h. Biofilm cultures were washed twice with sterile PBS and 1 ml of whole human blood from a healthy subject (pre-collected in EDTA tubes) was added to each biofilm well (Figs. 1 and 2). Well with only blood and no biofilm was used as negative control and incubated the stimulated sample plate in the incubator at 37°C in 5% CO2 in the air for 24 h.
After the incubation time point (24 h), the plates were removed from the incubator and whole blood stimulated biofilm, biofilm supernatant, planktonic and whole cells samples were collected in Eppendorf tubes. The sample tubes were immediately centrifuged at 5000 × for 5 min and collected the supernatants in tubes and stored at -20°C until the next step.
2.5. Cytokine Profiling using Membrane Arrays
A Human Cytokine Array Kit (Proteome ProfilerTM Antibody Arrays R&D SystemsTM), which can detect 36 human cytokines was used. Two ml of Array Buffer 4, which serves as block buffer, were added into each well of the 4-well multi-dish provided in the kit. Samples were prepared by adding 1 ml of each sample to 0.5 ml of Array Buffer 4 in separate tubes. To reach a final volume of 1.5 ml, Array Buffer 5 was used. 500 µL of the reconstituted Human Cytokine Array Detection Antibody Cocktail was added to and mixed with each prepared sample. The mixture was incubated for 1 hour. After blocking the array membrane to prevent non-specific binding, the samples were added onto the cytokine array membrane and incubated at room temperature for 1h. Following three washes in wash buffer, the array was treated with streptavidin HRP and incubated for 30 minutes at room temperature on a rocking platform shaker. The washed array was finally incubated with chemiluminescence reagent mixture and images were captured on SynGene G-box Imaging System. The positive signals seen on the array were identified by comparing it with the transparency overlay template with the pairs of reference spots in three corners of each array. SynGene tools analysis software was used to determine pixel densities (signal densities) of the spots on the array. The signal density of one of the reference spots with maximum intensity was graded 100 and used as a reference to calculate the mean spot pixel densities of the rest of the spots.
2.6. ELISA Quantification of Selected Cytokines
Based on the results from the cytokine array profile, absolute quantities of select cytokines (IL-8 and IL-10) were determined by ELISA. For this, ELISA immunoassay kits (Quantikine® ELISA R&D Systems) were used. A cytokine specific monoclonal antibody pre-coated ELISA plates pre-coated with cytokine-specific monoclonal antibodies were used in a quantitative sandwich enzyme immunoassay technique. To determine the absolute quantities of IL-8 and IL-10 in blood upon stimulation with biofilm, biofilm-supernatant, planktonic culture and whole cell preparations, the wells of ELISA plate were pipetted with standards and samples. Any unbound substances in the wells were removed with wash buffer and an enzyme-linked polyclonal antibody specific for cytokine of interest was added to the wells thereafter. Washing steps were followed to remove unbound antibody-enzyme reagent. Finally, a substrate solution was added to the wells and the color development was stopped. At the end of the assay, the intensity of the color developed in proportion to the amount of bound cytokine of interest was measured using iMarkTM Microplate Reader (Biorad).

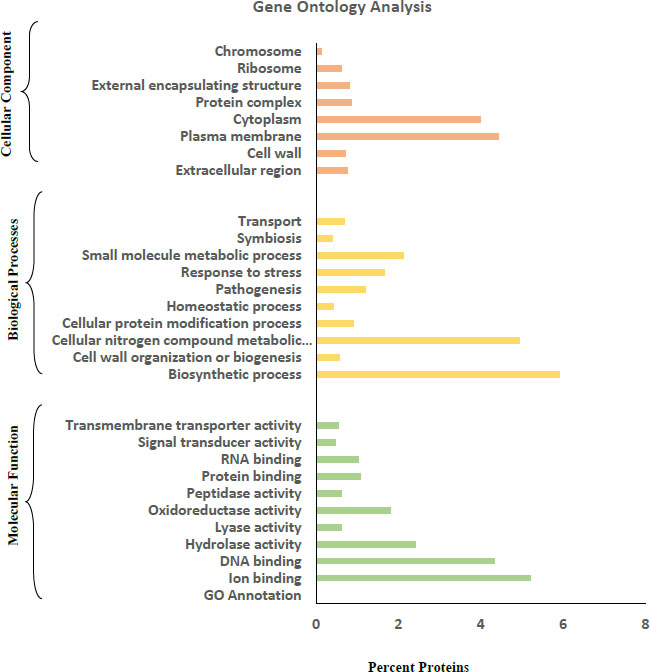
Blast2GO and the web software “CELLO2GO” were used for Gene Ontology annotation. Based on their features and functions, protein sequences were divided into three categories: Molecular Function, Biological Process, and Cellular Component.
3. RESULTS
3.1. In silico Analysis of R. dentocariosa Proteome
3.1.1. Proteins Secreted by R. dentocariosa
R. dentocariosa genome, which is about 2.4 million base pairs (53.8% GC content), encodes for 2116 proteins. The proteome contains 145 proteins secreted extracellularly as predicted by the bioinformatics tools SignalP and others. Of the 145 secreted proteins, 98 proteins were found to be secreted via “Sec” pathway while 47 proteins were identified to be secreted via “Twin-Arginine” (Tat) pathway. The number of lipoproteins secreted by this species was 41. Of the total 145 secreted proteins, current bioinformatics tools allowed us to specifically identify the names of only 33 proteins while the remaining were either hypothetical proteins or common Tat secretion pathway proteins (Table 1). When localization of the secreted proteins was analyzed (Fig. 1), 41.9% of the proteins were found to be released extracellularly, 18.6% anchored to the cell-wall, and 30% were membrane-bound. Interestingly, 9.3% of proteins were of cytoplasmic origin.
| Accession No. Protein Name |
|---|
| EFJ76380.1 actinobacterial surface-anchored domain protein |
| EFJ76381.1 anchored repeat ABC transporter, substrate-binding protein |
| EFJ76480.1 peptidase, S8 S53 family |
| EFJ76493.1 periplasmic binding protein |
| EFJ76532.1 ABC transporter, substrate-binding protein, QAT family |
| EFJ76545.1 ABC transporter, substrate-binding protein, family 5 |
| EFJ76547.1 ABC transporter, substrate-binding protein, family 5 |
| EFJ76556.1 LPXTG-motif cell wall anchor domain protein |
| EFJ76630.1 copper resistance protein CopC |
| EFJ76843.1 Transglycosylase-like domain protein |
| EFJ76876.1 peptidase, M23 family |
| EFJ76945.1 Imelysin |
| EFJ77006.1 penicillin-binding protein, transpeptidase domain protein |
| EFJ77086.1 thiol reductant ABC exporter, CydC subunit |
| EFJ77146.1 glycosyl hydrolase family 25 |
| EFJ77248.1 cytochrome c oxidase, subunit II |
| EFJ77253.1 menaquinol-cytochrome c reductase cytochrome c subunit |
| EFJ77339.1 export membrane protein SecD |
| EFJ77381.1 polysaccharide deacetylase |
| EFJ77382.1 excalibur domain protein |
| EFJ77406.1 LPXTG-motif cell wall anchor domain protein |
| EFJ77485.1 periplasmic binding protein |
| EFJ77513.1 ABC transporter, substrate-binding protein |
| EFJ77550.1 ABC transporter, substrate-binding protein |
| EFJ77829.1 molybdate ABC transporter, periplasmic molybdate-binding |
| EFJ77900.1 ABC transporter, substrate-binding protein, family 3 |
| EFJ77919.1 cytochrome c-type biogenesis protein CcsB |
| EFJ77922.1 redoxin family protein |
| EFJ78024.1 LPXTG-motif cell wall anchor domain protein |
| EFJ78045.1 NlpC P60 family protein |
| EFJ78188.1 Gram-positive signal peptide protein, YSIRK family |
| EFJ78214.1 conserved repeat protein |
| EFJ78263.1 iron manganese ABC transporter, periplasmic iron manganese-binding p |
3.1.2. Virulence Factors and Proteins with Potential Interactions with Host in the R. dentocariosa Proteome
The R. dentocariosa complete proteome was predicted to contain 698 proteins with virulence potential. The final prediction results were considered after deducing a consensus list based on Hidden-Markov Model (HMM) and Support Vector Machine (SVM) approaches using different tools. When only the secretome was used in the analysis, 18 proteins were predicted as potential virulence factors (Table 2).
| Accession No. Protein Name |
|---|
| EFJ76480.1 peptidase, S8/S53 family |
| EFJ76545.1 ABC transporter, substrate-binding protein, family 5 |
| EFJ76547.1 ABC transporter, substrate-binding protein, family 3 |
| EFJ77146.1 glycosyl hydrolase family 25 |
| EFJ77513.1 ABC transporter, substrate-binding protein, family 5 |
| EFJ77829.1 molybdate ABC transporter, periplasmic molybdate-binding protein |
| EFJ77900.1 ABC transporter, substrate-binding protein, family 3 |
| EFJ77922.1 redoxin family protein |
| EFJ78188.1 Gram-positive signal peptide protein, YSIRK family |
| EFJ76480.1 peptidase, S8 S53 family EFJ76630.1 copper resistance protein CopC EFJ77086.1 thiol reductant ABC exporter, CydC subunit EFJ77381.1 polysaccharide dssseacetylase EFJ77513.1 ABC transporter, substrate-binding protein EFJ77922.1 redoxin family protein EFJ78024.1 LPXTG-motif cell wall anchor domain protein EFJ78045.1 NlpC P60 family protein EFJ78263.1 iron manganese ABC transporter, periplasmic iron manganese-binding p |
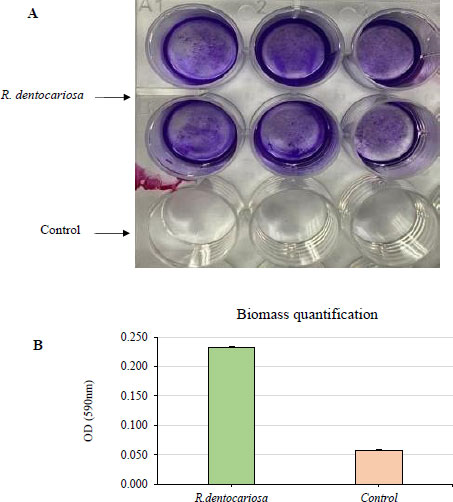
The biofilm biomass was stained (A) and quantified (B) by crystal violet staining after growing the biofilms for 2 days in 5% CO2 at 37 °C. The absorbance was measured at 590 nm wavelength.
3.2. Gene Ontology Analysis
Gene Ontology (GO) analysis of the amino acid FASTA sequences of the R. dentocariosa secretomes was achieved by using the tools Blast2GO and CELLO2GO (Fig. 2). In the GO category of molecular function, DNA binding (4.3%) and ion binding (5.2%) were the two major groups the proteins were annotated. A small percentage of proteins were also annotated to molecular functions such as transmembrane transporter activity, signal transducer activity and protein binding. Biosynthetic processes and cellular nitrogen compound metabolic processes were the major groups in the case of Biological Processes. Proteins involved in pathogenesis (1.2%), stress response (1.7%) and homeostatic process (0.42%) were also listed by the tools in GO annotation.
3.3. Biofilm Formation in R. dentocariosa
R. dentocariosa showed a moderate biofilm formation ability as shown in Fig. (2). Optical density values from crystal violet staining of the biofilms were between 0.2 and 0.3 absorbance units. This was also evident from the stereo microscopy images showing the stained biofilms.
3.4. Cytokine Profiling of Human Whole Blood Stimulated with R. dentocariosa
Cytokine antibody array analysis showed that the R. dentocariosa biofilm, biofilm-supernatant, planktonic, and whole cells, all induced CCL5/RANTES, ICAM-1/CD54, IL-8, macrophage inhibitory factor (MIF) and SERPIN-1 (Fig. 3). However, complement components, G-CSF, IL-2, and IL-16 were induced by only biofilm or planktonic cells. The whole cells failed to induce MIP-1α/MIP-1β, CXCL-1 and IL-6.
3.5. Absolute Quantification of Select Cytokines
Biofilm and planktonic cultures of R. dentocariosa induced the highest amounts of IL-8, i.e., ~2000 pg/ml (Fig. 4). The biofilm-supernatant induced a mean (SD) value of 1432 (104) pg/ml, while the whole cells induced 285 (26) pg/ml. In the case of IL-10, planktonic cultures induced significantly higher cytokine levels (296 pg/ml) (P<0.05) followed by biofilms, biofilm-supernatants and whole cells that induced IL-10 between 61 (7) and 96 (11) pg/ml (Fig. 5).

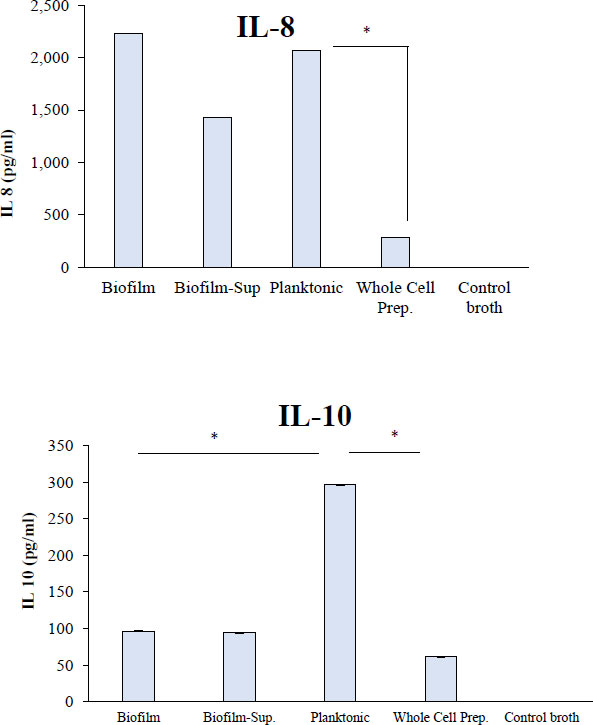
. Human whole blood was treated with R. dentocariosa biofilms, biofilm-supernatants, planktonic cells and whole cells. After the stimulation period, plasma was separated from blood and the cytokines were quantified by using standard ELISA Kits for specific cytokines *P<0.05.
4. DISCUSSION
In silico analysis of the proteome of R. dentocariosa revealed numerous proteins that are potentially secreted and that possess virulence properties. Interestingly, the analyses further showed that a large number of proteins (n=47) are secreted via the Tat pathway. The species also contains about 40 lipoproteins. Several of the proteins predicted to be virulence factors were found to be well-established virulence factors in other pathogenic bacterial species (13). Gene Ontology analysis was performed on the FASTA sequences to get insight into the functional importance of the proteins identified from the R. dentocariosa proteome. With the biosynthetic process, cellular nitrogen compound metabolic process, and small molecule metabolic process, the secretome had a higher percentage of proteins in the “biological processes” category. A higher number of proteins with protein binding, DNA binding, and ion binding activities were found in the secretome. Despite the limitation that we did not experimentally analyze the protein profiles of the R. dentocariosa secretome from biofilm and planktonic cells, the data on predicted virulence factors and secreted proteins may correlate well with the immunostimulatory potential of this species.
In the present study, biofilms and planktonic cells appeared to have different cytokine stimulating potentials. Our ELISA results showed that while the planktonic cells induced significantly higher IL-10 quantities, the IL-8 was induced at similar levels both by biofilm and planktonic cells. Cytokine membrane profiler assays revealed stimulant-dependent differences in cytokine induction. In general, R. dentocariosa showed differences in cytokine stimulating potential when different stimulants such as biofilm, planktonic, biofilm-supernatant and whole cells were used. It is conceivable that the host immune response could influence the various results related to the type of stimulant. Several earlier studies have made similar observations that planktonic cells induced higher amounts of the cytokine than biofilms [15-17]. Despite the current understanding that biofilms possess enhanced virulence potential in terms of surface adherence, increased antibiotic resistance and increased survival in the host, they induce a lower amount of cytokines perhaps as a survival strategy.
Our study found that R. dentocariosa forms a moderate biofilm in comparison to other strong biofilm-forming oral species [14]. The ability of R. dentocariosa to form biofilm alone or in the presence of other oral microorganisms has been demonstrated previously [18, 19]. The biofilm potential of R. dentocariosa in endocarditis is largely unknown; however, Greve D et al. [20] found that the species was indeed present in the specimen from heart valves.
Earlier studies have indicated that R. dentocariosa is associated with periodontal inflammatory disease [21, 22]. However, little is known about the pathogenicity of this bacterium. To characterize the host response to this bacterium, Kataoka et al. 2014 [7] measured (using ELISA) the amount of TNF-α in the culture supernatant following the stimulation of THP-1 cells (a human acute monocytic leukemia cell line) with R. dentocariosa cells and reported that this species induces host TNF-α production by a TLR2-dependent mechanism [7].
The potential of R. dentocariosa in inducing cytokines such as TNF-α, IL-1β, IL-8, IL-10 etc. has been reported earlier; however, the target cells were different in those studies, i.e., THP-1, peripheral blood lymphocytes, etc. Induction of more soluble interleukin (IL)-6 receptor a (sIL-6Ra), sIL-6Rb and tumor necrosis factor ligand superfamily (TNFSF)13B from OKF6 keratinocytes by Primary Biliary Cholangitis (PBC) salivary microbes, and induction of more IL-6, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), chemokine (C-C motif) ligand (CCL)13, C-X-C motif chemokine (CXC)L1 and CXCL16 by PBC salivary supernatant from THP-1 macrophages was observed in comparison to healthy controls [23]. Compared with healthy controls, Rothia along with Enterococcaceae, Granulicatella, and Streptococcus was noticed to be among the depleted taxa in PBC saliva while Bacteroidetes, Campylobacter, Prevotella and Veillonella were among the enriched taxa [23]. In another study, when crude cell wall and cytoplasmic antigens derived from R. dentocariosa were applied to peripheral blood lymphocytes and subjected to radioactive thymidine; the resulting lymphocyte blastogenesis was surveyed. All three groups displayed statistically similar levels of stimulation (F = 0.71), demonstrating that crude antigens of Rothia dentocariosa are not appreciably active in vitro studies of cell-mediated immunity, as measured by lymphocyte blastogenesis [24]. Further, R. dentocariosa potentiates the production of other important cytokines such as interferons by the peritoneal cells of BALB/c mice [25].
A major limitation of this study is that using a multiplex biomarker quantification assay, a greater number of proinflammatory cytokines could have been studied upon stimulation with different culture preparations of R. dentocariosa. This is primarily because of the limited resources that were available for this study.
CONCLUSION
Identification of potential virulence factors predicted to be secreted extracellularly may suggest a key role for R. dentocariosa in oral and non-oral infections but requires experimental validation using mechanistic studies. In general, R. dentocariosa showed differences in cytokine stimulating potential when different stimulants such as biofilm, planktonic, biofilm-supernatant and whole cells were used. This may suggest differences in the composition/concentration of the bacterial components in the stimulants, with varying abilities to induce cytokine production, maybe the reason for the observed differences.
LIST OF ABBREVIATIONS
| OD600 | = Optical Density |
| WCP | = Whole Cell Preparations |
ETHICS APPROVAL AND CONSENT FOR PUBLICATION
The Kuwait University Health Sciences Center's Ethical Committee approved this study (DR/EC/3413).
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
The blood donor was given written information about the study's nature and aims, and when the volunteer agreed to participate, written informed consent was obtained.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
We thank the Research Administration of Kuwait University for generous funding to the Oral Microbiology Research Laboratory (KU Grant SRUL 01/14).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Research Administration of Kuwait University for generous funding to the Oral Microbiology Research Laboratory (KU Grant SRUL 01/14).


