Severe Acute Respiratory Coronavirus-2: A Critical Review of Virus Biology, Genome and Pathophysiology
Abstract
Severe Acute Respiratory Coronavirus-2 [SARS-CoV-2] emerged as a great threat to the world at the end of December 2019 in China. The SARS-CoV-2 evolved from a virus responsible for the SARS epidemic in 2002. The SARS-CoV-2 has a high rate of human-human transmission and originated from the bat. It has a close resemblance with bat-like-SARS-CoV compared to SARS-CoV; however, the Spike protein responsible for virus-host cell interaction possesses the least similarity with that of SARS-CoV. Cytokine Storm is associated with the severity of Covid-19 and leads to acute respiratory distress syndrome [ARDS] and/or multiple organ dysfunction syndromes [MODS]. In the current review article, the features of a novel coronavirus, including viral biology, genomic organisation, life cycle, pathophysiology and genetic diversity, have been discussed. The development of policies and plans which can prepare the world for future pandemics has also been proposed. In addition, the drug development pipelines, diagnostic facilities and management of such pandemics need an up-gradation to contain the current as well as future outbreaks.
1. BACKGROUND
The world had just coped with the epidemic of MERS-CoV, which caused respiratory distress syndrome in the middle-east in the year 2012-2016; the human race has to face the outbreak of a novel coronavirus pandemic again [1]. In December 2019, Wuhan, a city in China, reported a plethora of inexplicable cases of pneumonia that, in time, spread abruptly within China and other parts of the world. After a few days, the pathogenic agent responsible for the unexplained respiratory disease was identified as a new strain of coronavirus called SARS-Cov-2 [2]. Realizing the extensive outbreak of the virus, WHO, on 30th January 2020, declared the coronavirus epidemic as, “public health emergency of international concern”. Coronaviruses (CoVs) which constitute the family of Coronaviridae, are large, enveloped viruses possessing a positive-sense single-stranded RNA. The viruses that infect both animals and humans are divided into four genera depending upon their genotype and serotype as Alpha CoVs, Beta CoVs, Gamma CoVs and Delta CoVs. Hitherto, the coronaviruses, which infect the human race, belong to the first two groups. Howbeit, the Beta CoVs have gained considerable attraction across the world, causing two epidemics and one pandemic, respectively [3, 4]. The first outbreak of coronavirus occurred in the year 2002-2003, people suffered from Severe Respiratory Distress Syndrome (SARS), and China was the epicentre of this epidemic. The SARS-CoV affected 26 countries worldwide, infecting more than 8000 with a death toll of approximately 800 patients. Another virus of the same genre known as MERS-CoV came into existence in 2012. According to the recent updates in 2019, around 2494 infected cases with 858 deaths have been reported across 27 countries in the world [5]. Unfortunately, the SARS-CoV-19, also knows as Novel Coronavirus, Covid-19 is far more aggressive than the two viruses affecting above 23 million people around the globe with > 8 lakh fatality cases as of 25th August 2020 [6, 7]. These viruses are zoonotic agents originating from animals and causing infection in humans. Thus identifying the evolution cycle of Beta CoVs genera according to their structure and function is the thrust area of today’s time. In addition, due to the unavailability of a specific treatment modality for Covid-19 disease, the current review will help understand the perspectives of treatment pipelines. Therefore, the current article presents an overview of the virus biology, pathophysiology, the importance of saliva as a diagnostic indicator, and the various treatment modalities in the pipeline.
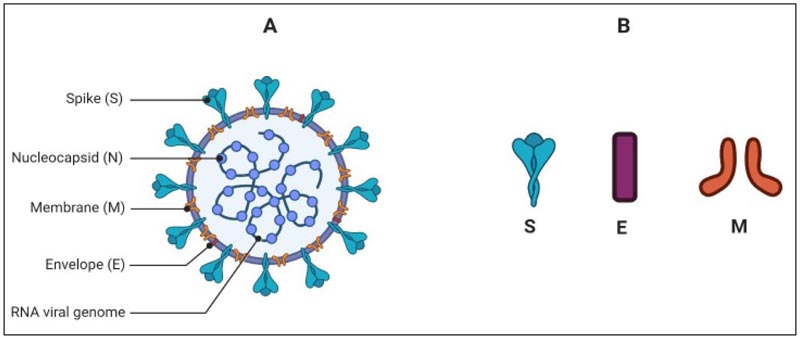
2. VIRUS MORPHOLOGY
Coronavirus is a spherical and pleomorphic particle whose diameter may range from 65-120 nm. The genome of the virus is enveloped by lipid bilayer from where it projects the pear-like, club-shaped spike glycoproteins (Fig. 1). The spikes taper towards lipid envelops with an average 10nm width distally. Some of the other strains of coronaviruses may contain additional small projections of size 5-10 nm of Hemagglutinin-Esterase (HE) protein [8]. There are four main structural proteins of the virus; spike protein (S), envelope small membrane protein (E), membrane protein (M) and nucleoprotein (N). Out of these four proteins, the first three are the membrane proteins, and the S glycoprotein is pivotal as it guides the virus-host interaction. The S protein consists of approximately 1200 amino acid (AA) chain and belongs to the class-I fusion proteins. S protein of the SARS-CoV-2 is known for its key functions like receptor binding, host response and pathophysiology [9-11]. The M and E proteins are crucial for giving a proper shape to the virus, its assembly and the viral pathophysiology. The M protein was the first viral membrane protein to be described and was formerly known as the E1 protein
M protein has three transmembrane hydrophobic domains and also has a large amphiphilic carboxy-terminus domain located in the cytoplasm, which interacts with the nucleocapsid once the virus enters into the host cell [12, 14]. E is a small polypeptide, which is profusely expressed inside the target cell, however, the bilayer lipid membrane incorporates only a small portion of E protein. It helps in the assembly and budding of the CoV [15]. The N protein binds with the virion genome in a beads-on-a-string fashion to form the nucleocapsid. Besides this function, N protein has played its role in virus replication and the host response. In the replication cycle [discussed later], the protein functions in assembling the membrane proteins and the nucleocapsid to form virus bud [16]. In addition, some studies reported that this protein can bind with nsp 13 to secure the RNA to replication-transcription complex (RTC) [17, 18].
2.1. Genomic Organisation of SARS-CoV-2
The genome of SARS-CoV-2 possesses a single-stranded positive-sense RNA (+ssRNA) encapsulated by nucleocapsid protein N [19, 20]. The single-stranded genome is ~29.9 kb (2891 base pairs) in size with 5′cap and 3’ poly(A) tail and may range from 26-32 kb, which encodes about 9860 amino acids (AA) [19, 21]. The G+C contents range from 32%-43%. The coronaviruses are considered to contain the largest RNA in nature as virtually all the viruses holding RNA genome are three to four times smaller than the coronavirus genome [22, 23]. The RNA encloses variable numbers of open reading frames (orfs) with at least six orfs and may extend up to twelve, which are bounded by untranslated regions (UTRs) [24, 25]. About two-thirds (67%) of the complete virus genome consist of two ORFs; ORF1a/b, which serves as the template to translate the replicase polyproteins 1a/1ab (pp1a/1ab) [25]. Ribosomal frameshift site interconnects ORFs 1a/b and at this site occurs the processing of the replicase gene in a -1 frameshift between the two ORFs resulting in the synthetisation of pp1a and pp1b [21]. Further, these proteins are processed by the virally encoded 3C-Like Proteinase (3CLpro), also called the chymotrypsin-like and papain-like proteinase (PLpro), to release the sixteen non-structural proteins or nsps [26-28] (Fig. 2). The remaining ORFs occupies the remaining one-third of the genome encoding the four structural proteins and other accessory proteins of the virus (Fig. 2).
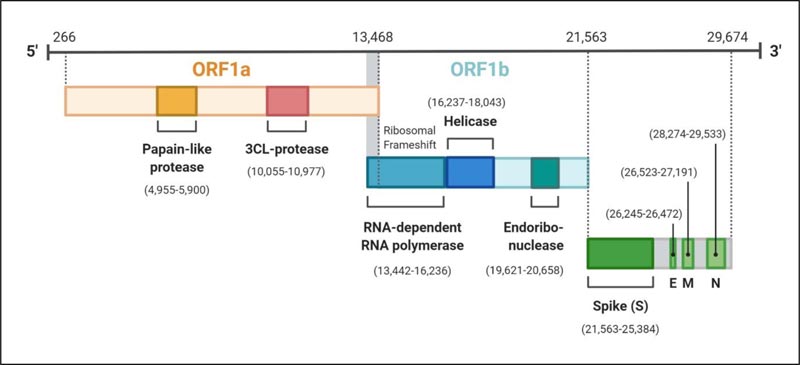
The four structural proteins are invariably arranged in the order S-E-M-N near the 3’poly A tail [29]. The structural and accessory proteins are translated by the sub-genomic RNA (sgRNA) [30]. The extra-genes (ORFs), which translate some special and accessory proteins of the virus, are scattered among the canonical genes of the coronaviruses. These proteins, like HE protein, 3a/b protein, and 4a/b protein, vary from one genus to another in the family Coronaviridae [22, 31]. The accessory proteins of the coronaviruses are least studied; however, they are considered to be crucial for virus replicated and also to counteract the host response [31, 32]
3. LIFE CYCLE OF SARS-CoV-2
3.1. Receptor Recognition Mechanism and Virus Entry
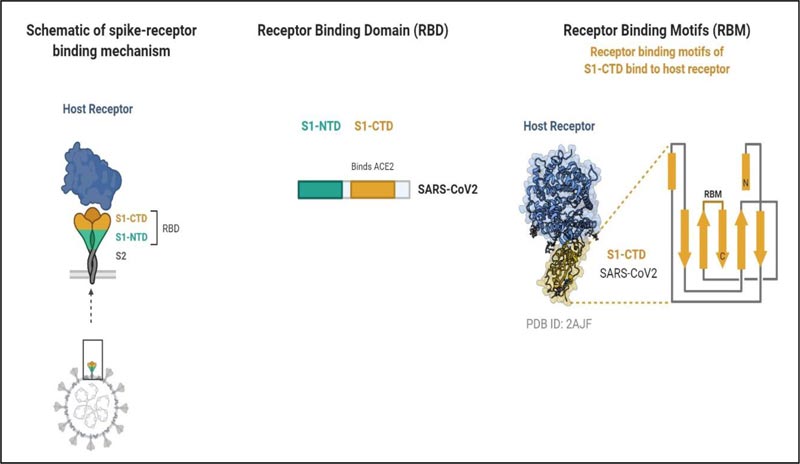
The SARS-CoV-2 entry into the target cell is governed by the spike (S) protein, and it recognizes the human Angiotensinogen Converting Enzyme-2 (hACE-2) receptors to enter into the host similar to SARS-Cov [33, 34]. S protein is a transmembrane glycosylated protein, which forms homotrimers projecting from the viral membrane in a spike-like fashion. The S protein is functionally categorized into two domains or subunits responsible for binding host cell receptor (S1 domain) and fusion of the cell and virion membrane (S2 domain). The S2 subunit is more stable than the S1 subunit [11]. S1 subunit comprising Receptor-Binding Domains (RBDs) initiates the virus-host cell interaction. Further, the RBDs possess a core structure and Receptor-Binding Motif (RBM). It is this RBM that attaches to the ACE-2 [34] (Fig. 3). Before receptor binding, S proteins exist in a stable prefusion trimer undergoes conformational change following the binding of the receptor-Binding Domains (RBDs) to the receptor. This results in the deterioration of the S1 and a shift to a stable conformation of S2 subunit.
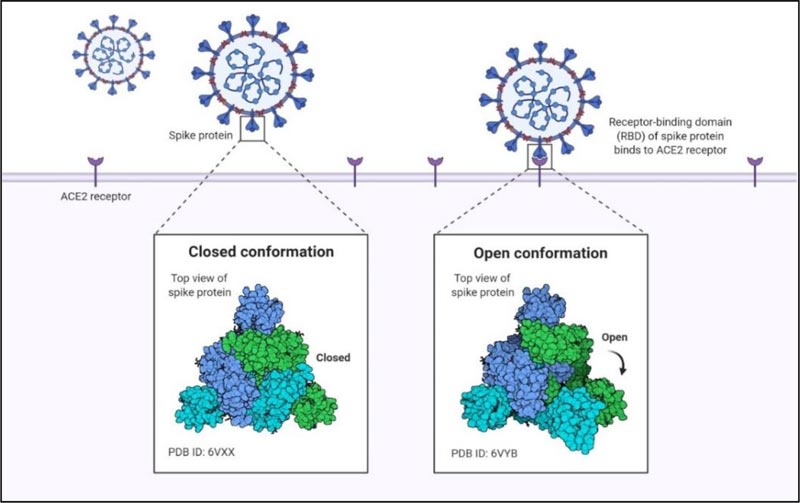
The Spike protein exists in different conformational states like open conformation and close conformation. During close confirmation, the receptor binding determinants of the RBDs are unable to recognize hACE-2 while they can easily recognize and attach to the target cell receptor during the open conformation (Fig. 4). These various conformations and the dense glycan guard the virus against the host’s immune response. Following the initial attachment, S2 subunit functions to fuse the viral and host cell membrane.
The cleavage of S proteins of SARS-CoV and MERS-CoV mostly occurs at the S1/S2 site. Some studies show that proteases like TMPRSS2 and cathepsin L cleaves the S protein of SARS-CoV at the S1/S2 and S2’ regions, and the process is called priming [35, 36]. However, one study reported a unique priming site in SARS-CoV-2 called the S1/S2 furin cleavage site responsible for the initial binding with the receptor. This was named after the sequence analysis of the virus S protein, which showed four amino acid residue insertion at the S1/S2 border when compared with the SARS-Cov [33]. The second proteolytic cleavage occurs at the S2’ site. Thus, priming occurs at the S1/S2 and S2’ site for mediating virus entry into the cell. Fusion occurs in the presence of key components like fusion peptides; Internal Fusion Peptides (IFPs), Fusion Proteins (FP) and Heptad Repeat one and two (HR1 and HR2). These domains drag the viral envelope in proximity to the target cell membrane. In addition to this, host/human proteases are crucial for fusion to occur [37].
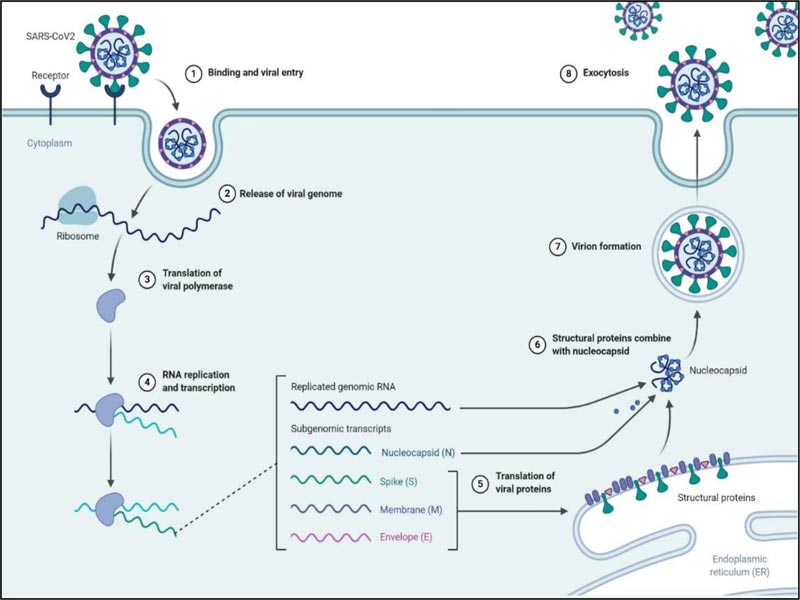
3.2. Replication
After entering into the host cell via endocytosis, the viral genome is released in the cytoplasm, and the RTC is formed by the 16 nsps in double-membrane vesicles [38]. One of the main proteins of RTC RNA-dependent RNA polymerase initiates the replication of the +ssRNA to form sets of minus subgenomic RNAs (-sgRNA) employing discontinuous transcription. The formed nested sets minus subgenomic RNAs serves as the template to form subgenomic messenger RNAs (mRNAs). Besides RNA-dependent RNA-polymerase, the RTC contains other proteins like helicase, endoribonuclease and exoribonuclease, which assist in this complex process (Fig. 2) [25, 39]. Unlike other RNA viruses, the exoribonuclease enzyme among the 16nsps is unique to coronaviruses, which functions to proofread the RTC [40, 41]. The subgenomic mRNAs are used to translate the structural proteins [S, E, M] of the virus in the endoplasmic reticulum and subsequently, they travel to the ER-Golgi intermediate compartment [ERGIC] where the N protein is formed. The +ssRNA of the virus is replicated parallel to the translation process. (Fig. 5) [15, 24, 33].
3.3. Release
Budding occurs at the ERGIC through the assembly of the proteins and RNA, and the mature virus-like particle is formed, which is then fuses to the inner membrane, and the virus is released outside to infect the cells [38].
Fig. (5) depicts the complete replication cycle of SARS-CoV-2 inside the host cell.
4. IMMUNOPATHOGENESIS MECHANISM OF SARS-CoV-2
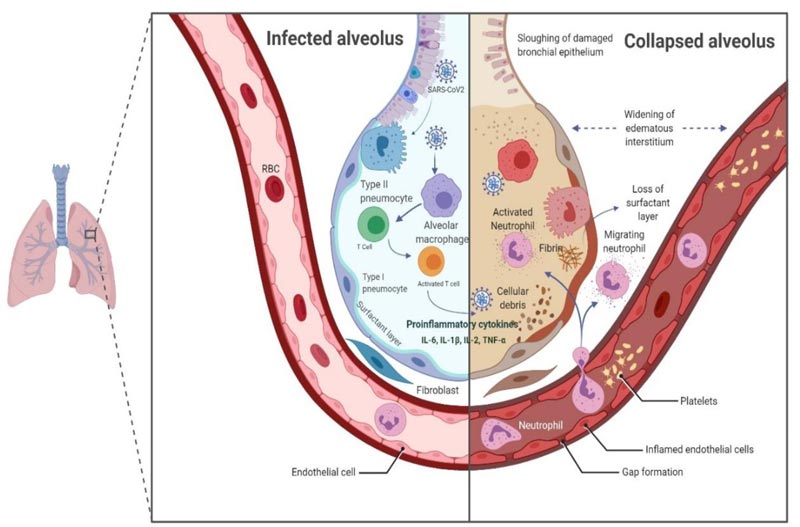
The Type II alveolar epithelial cells are the chief targets of Covid-19 infection, and they choreograph the exacerbation of pro-inflammatory mediators. SAS-CoV2 replicates inside these cells, and subsequently, several viral copies are delivered into the alveolus, thereby leading to apoptosis of Type II cells. The Pattern Recognition Receptors (PRRs), such as toll-like receptor (TLR), RIG-I-like receptor (RLR), NOD-like receptor (NLR) and C-type lectin-like receptors (CLmin) identify pathogen-associated molecular patterns (PAMPs) of the virus and provoke an inflammatory response which engages the macrophages at the site of cell injury [42, 43]. The macrophages further draw other immune cells like T lymphocytes, including CD4* and CD8* cells. CD4* activate B cells to produce antigen-specific antibodies that neutralize the viruses in the alveolus, while CD8* T cells have a cytotoxic effect on the virus, reducing the infection. Inadequate viral clearance and damage to lymphocytes hyper-activates the T cells instigating an increase in the production of cytokines and chemokines like IL-1β, IL-2, IL-6, IL-7, IL-8, tumour necrosis factor-α [TNF-α], Interferon-g and chemokines like C-C motif chemokine ligand (CCL)-2, CCL-3, and CCL-5 [44, 45]. This dysregulation in the immune response, the “cytokine storm” is virtuously exerted by the cell-mediated immune response (Fig. 6). An elevated cytokines and chemokines profiles in the pulmonary tissue and peripheral blood suggests that these factors play a vital role in the immunopathogenesis of SARS-CoV-2. There is a strong association of the disease severity with elevated serum levels of IL-6. Critically ill patients have higher IL-6 levels compared with moderately and ordinary ill patients [46-48]. The cytokine storm is pivotal in modulating the clinical outcome of the patients. It marks the disease severity and is responsible for Acute Respiratory Distress Syndrome (ARDS) and/or Multiple Organ Dysfunction Syndrome (MODS) in Covid-19 patients.
The association of Covid-19 disease with deranged coagulation parameters and connection between DIC (disseminated intravascular coagulation) and Covid-19 have led to the search for the effectiveness of anticoagulant molecules as supportive therapy in SARS-CoV-2 positive patients [49]. The coagulopathy, mainly in the form of venous thromboembolism [pulmonary embolism], is often correlated with the poor prognosis [50]. The shreds of evidence supporting this theory derive from case reports and studies depicting a significant increase in D-dimer and/or fibrin degradation products (FDPs) of severe Covid-19 patients [51-54]. In another study, a better prognosis was observed in patients who had received low molecular weight heparin as anticoagulant therapy [55]. This is an emerging concern and requires more investigations to arrive at a decision on whether anticoagulant therapy should be administered prophylactically.
Laboratory analysis of SARS-CoV-2 positive patients also reveals increased levels of ferritin, C-reactive protein (CRP) and procalcitonin [47, 56]. Elevated ferritin levels are a sign of bacterial and viral contamination, and it is evident that hyperferritinemia leads to activation of macrophages, which further releases cytokines into the blood, thereby contributing to the development of cytokine storm in Covid-19 patients [57]. The hypoxic environment is one of the factors responsible for hyperactive inflammation and disease severity. This is because SARS-CoV-2 attacks the β-chain of haemoglobin, which dissociates iron and form porphyrin in the body. Ultimately, the oxygen-carrying capacity of the blood reduces, giving rise to the hypoxic condition. This condition may worsen in pre-existing respiratory illness [58].
5. GENOMIC DIVERSITY OF SARS-CoV-2
The causative agent responsible for the atypical pneumonia outbreak was a new strain of the virus, which fell under the β coronavirus. Initially, the virus was named as 'WH-Human 1' coronavirus in a study for the patient who was admitted on 25th December 2019 [3]. Subsequently, other studies proved that 'WH-Human 1' coronavirus, which is now called 2019-nCoV or SARS-2, belongs to the Sarbecovirus subgenus of β coronavirus genera, along with SARS-CoV. MERS-CoV is a member of the same subgenus (Merbecovirus) [3, 19, 59]. SARS-CoV-2 closely relates to the three bat-SAR like-CoronaVirus (bat-SL-CoV), bat-SL-CoV-RaTG13, bat-SL-CoVZC45, and bat-SL-CoVZXC21. Bat-SL-CoV-RaTG13 has the highest sequence identity with SARS-CoV-2 of 96.3%, while the other two have 87·99% and 87·23% sequence identity, respectively [19, 59]. The sequence identity of the SARS-CoV (~79%) and MERS-CoV (~50) is less when compared with bat-SL-CoV [8, 19]. The genomic organisation of SARS-CoV-2 bears a resemblance to the gene structure of other β coronaviruses, and it is very close to the SARS-CoV. The longest ORF1ab that contains the 16 nsps and other downstream ORFs is also similar to the SARS-CoV. However, the genomic divergence is noted in the S protein of SARS-CoV-2 from the SARS-CoV [60]. As discussed earlier, the insertion of the furin cleavage site at the S1/S2 junction has contributed to the diversity and increase pathogenicity of the virus. Departure from the genomic identity of the spike protein of SARS-CoV-2 from SARS-CoV and MERS-CoV reveals that the trajectory of SARS-CoV-2 is different and more aggressive from these viruses [29, 33, 61]. The close resemblance of the bat-like coronavirus and the SARS-CoV-2 made it clear that the bats are the natural reservoir of the virus. However, a study reported pangolin as a reservoir of the virus [19, 20]. There is evidence that suggests the presence of an unknown intermediate host between the bat to human transmission of the SARS-CoV-2 similar to SARS-CoV (civets) and MERS-CoV (camels) [62].
6. SALIVA: A DIAGNOSTIC WINDOW TO COVID-19
Saliva is a complex biological fluid comprising of water, salts, metabolites, molecules, proteins as well as a plethora of microbes, including bacteria and viruses, responsible for various pathologies [63, 64]. Regardless of the heterogeneous nature of saliva as it contains a wide spectrum of secretions like gingival crevicular fluid, glandular fluids, oral exfoliated cells, etc., it has been demonstrated as a good diagnostic indicator for oral and systemic diseases [65]. Further, the viruses like Hepatitis, Ebola, Zika, SARS-CoV, and MERS-CoV have been detected in saliva samples of patients, making saliva a potent diagnostic tool for the detection of viral infections [66]. In the COVID-19 scenario, the most reliable test for specific and accurate identification of SARS-CoV-2 infection is the Reverse Transcription Polymerase Chain Reaction (RT-PCR). The upper respiratory specimens collected to determine the virus RNA are in the forms of nasal swabs, nasopharyngeal or oropharyngeal swabs, mid-turbinate swabs. The lower respiratory specimens may be in the form of bronchoalveolar lavage, nasopharyngeal aspirate, sputum in case of patients reporting with pneumonia [67]. Howbeit, several studies have prompted the non-invasive nature of the sample collection resulting in patient haemorrhage and discomfort. Besides, there also exists a risk of virus transmission to the health care workers collecting the samples [67-69]. Saliva samples have astounded these undesirable outcomes and proved to be an effective non-invasive specimen for laboratory detection of COVID-19. The relative ease of saliva collection as well as minimal or no risk of virus contamination to the health-care personnel mark saliva as a potent biological specimen for disease diagnosis [70, 71]. However, a study has observed an enhanced efficiency of virus detection from the broncho-alveolar lavage samples, especially in the severely ill SARS-CoV-2 infected cases [72]. Considering that each technique or method possesses certain merits or demerits, saliva can be a reliable alternative diagnostic tool in the COVID-19 pandemic.
7. THERAPEUTIC MODALITIES AGAINST SARS-CoV-2
In the current scenario, no specific single or combined antiviral drug therapy exists and the main treatment plan is observation and supportive care. The recombinant IFN in combination with remdesivir, had shown promising results to decrease the virus load [38, 73]. Chloroquine has also shown favourable results to some extent by inhibiting the steps involved in the virus replication or down streaming certain immune markers like TNF-α and IL-6 [74, 75]. Chloroquine concomitantly with remdesivir, depicted inhibitory results in-vitro [76]. Quite a few individuals are investigating the effectiveness of convalescent plasma in reducing the mortality of severe Covid-19 patients. Convalescent sera therapy is adoptive immunotherapy in which neutralizing antibodies from the recovered Covid-19 patients are administered to the patients as an adjunct to the standard treatment. Significant clinical outcomes have been achieved with this therapy, along with declination of the virus load [77-79]. As discussed earlier, elevated levels of IL-6 is present in severely ill SARS-CoV-2 positive patients; some groups have evaluated tocilizumab, an antagonist of the IL-6 receptor. The preliminary results showed promising effects of tocilizumab like a decrease in the IL-6 level, clinical and radiographic improvement and also reduced mortalities [80, 81]. To date, no specific or particle therapeutics are existing against Covid-19, and many approaches [mentioned above and other] have been investigated for their effectiveness, however, more studies are desirable to endorse their safety and efficacy.
CONCLUSION
Coronaviruses are identified in several species around the earth, and a plethora of these is yet to be discovered. Recently, a group of researchers discovered seven strains of coronaviruses from the bat in Myanmar, out of which six are the newly detected strains [82]. There is still a broad spectrum of scope to understand the genome of the virus and its pathophysiology at the molecular level. The certainty of virus mutations and its asymptomatic and mild behaviour in humans has emerged as a huge burden on healthcare systems across the globe. To date, there is deprivation of a specific therapy or vaccine against Covid-19. The Discovery of viable drug therapeutics would substantially alter the course of the Covid-19 outbreak aiding the eradication of the virus, thus saving lives on this planet. The virus can be contained following social distancing and personal sanitation. Policy discussion, proper vigilance and surveillance should be carried out to prevent future outbreaks of such deadly diseases.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
This paper would not have been possible without the exceptional support of my institute Datta Meghe Institute of Medical Sciences. I am also grateful for the insightful comments offered by the anonymous peer reviewers at The Open Dentistry Journal. The figures in the manuscript were prepared using “Biorender” software.


