All published articles of this journal are available on ScienceDirect.
Metagenomic Assessment of Different Interventions for Treatment of Chronic Periodontitis: A Systematic Review and Meta-Analysis
Abstract
Background:
Chronic periodontitis is attributed to oral microbial imbalance and host inflammatory reaction.
Objective:
Our review addresses the question of: Are the available interventions able to regain oral microbial balance in patients having chronic periodontitis?
Data Sources:
We performed a comprehensive systematic search of MEDLine via Pubmed, Cochrane CENTRAL, Clinicalkey, Clarivate Analytics, Springer materials, Wiley, SAGE, Elsevier, Taylor & Francis group, and Wolter Kluwer, together with hand searching and searching the grey literature.
Eligibility Criteria:
We included interventional studies testing the microbiome analysis using metagenomic techniques as an outcome to any intervention for chronic periodontitis.
Study Appraisal and Synthesis Methods:
All studies were imported in Mendeley. The risk of bias was assessed using the specific tool for each study design. The results were analysed using RevMan. All the review steps were performed in duplicates.
Results:
The search yielded 2700 records. After exclusion steps, 10 records were found eligible. We included 5 RCTs, 1 non-RCT, 3 before-and-after studies, and 1 ongoing study. The studies tested non-surgical periodontal treatment with and without antibiotic coverage, probiotics, sodium hypochlorite rinse, and different toothpaste ingredients. One RCT tested the use of enamel matrix derivatives in cases with furcation involvement.
Limitations:
The eligible available studies were small in number. Also, the risk of bias and lack of a standardized protocol impaired the ability to pool all the results.
Conclusions:
The body of the available evidence is not sufficient, and future studies are recommended to better evaluate the effect of periodontal treatments on the periodontal microbiome.
1. INTRODUCTION
Periodontitis has been proven to be a chronic multifactorial disease. The progressive damage of tooth-supporting structures occurring in periodontitis is attributed to host-mediated inflammation and oral microbial imbalance [1, 2].
The damage process of periodontitis is known to be primarily ignited by gingival inflammation.
Both the gingival tissue breakdown and the inflammatory products enrich certain species of the oral microbiome. These dysbiotic changes then trigger the host cells to produce proteinases that mediate loss of marginal periodontal ligaments, apical migration of the junctional epithelium, and apical spread of bacterial biofilm [2].
Being a cornerstone in the etiopathogenesis of periodontitis, the oral microbiome dysbiotic changes have represented a primary concern of scientific research for decades. The research aimed to detect the exact mechanism of dysbiosis so that clear pathophysiology of the disease can be clarified. Thus, a specific intervention targeting the exact dysbiotic changes can be implemented to ensure effective treatment and better prognosis [3].
Aiming to detect the oral microbiome, research used various methods starting by growing certain bacterial culture on certain media, detecting certain strains with known Nucleic Acid (NA) sequences by NA hybridization, amplifying certain strains of bacteria and virus with known NA sequences by Polymerase chain reaction, and ending up with performing NA sequencing to all the microbiome and by doing so, detecting all the known and unknown strains [4].
The NA sequencing methods are characterized by being specific to the microbial NA by sequencing the unique microbial 16S ribosomal RNA. 16S rRNA sequencing can be performed by different sequencing technologies including shotgun, pyrosequencing, and the lately evolved, Next-generation – also called high throughput- sequencing. All these technologies are referred to as metagenomic sequencing [4, 5].
Thus, metagenomic sequencing would count as the most reliable existing method to detect the ability of the treatments of periodontitis to target the pathologic dysbiotic changes regaining the microbiome balance; and so, test the effectiveness of the treatment [6].
Therefore, our review aims to assess the effectiveness of different interventions used for the treatment of chronic periodontitis assessed by metagenomics methods. The review questions the ability of the interventions to regain oral microbial balance in patients having chronic periodontitis.
2. MATERIALS AND METHODS
2.1. Eligibility Criteria
The protocol and the full report of this systematic review followed the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines.
All Randomized controlled clinical trials (RCTs), Non-Randomized controlled trials (non-RCTs), and Before-and-After studies which included any type of intervention to treat patients with chronic periodontitis were included in our review. When an eligible study was found to include only one arm that assesses an intervention for the treatment of periodontitis, this arm only was included in the review as a before-and-after study. The same protocol was followed with ongoing studies as well.
A study was considered eligible when it includes adult, systemically healthy, non-smoker patients diagnosed clinically with chronic periodontitis. The primary outcome for our review is the microbiome diversity index measured after performing the metagenomics analysis of the supragingival or subgingival samples. Studies including only saliva samples were excluded. Our secondary outcomes include the clinical presentation of the disease assessed by bleeding on probing, clinical attachment loss, and/or probing depth.
The search had no language restrictions; however, time was restricted from the year 2005 to date as metagenomics technology was not introduced earlier.
2.2. Information Sources and Search
A systematic search was performed using the synonyms of periodontitis, plaque, intervention, and microbiome in MED Line via Pubmed and Cochrane Controlled Register of Trials (CENTRAL). Clinicalkey, Clarivate Analytics, Springer materials, Wiley, SAGE, Elsevier, Taylor & Francis group, and Wolter Kluwer were all searched through the Egyptian Knowledge Bank search engine (ekb.eg). Reference lists of the included trials and grey literature search on OpenGrey were searched.
The following search strategy was followed: ((((((((((((((((treatment) OR therapy) OR debridement) OR scaling) OR root planning) OR brushing) OR brush) OR tooth paste) OR paste) OR antibiotic) OR probiotic) OR mouthwash)) OR antimicrobial)) AND (((((((((microbiome) OR microbiomic) OR metagenome) OR metagenomic) OR 16S rRNA) OR 16S rRNA sequenc$)) AND ((((((oral) OR mouth) OR gums) OR gingiva) OR plaque) OR dental plaque)) AND ((((periodontal) OR periodontitis) OR periodontal pocket) OR pocket))).
2.3. Study Selection
The search for studies started in October 2018 and ended in May 2019. All studies that were judged as eligible from the title and abstract or the eligibility was not confirmed at this step were then read in full text. The included studies were then de-duplicated using Mendeley (Version 1.17.10) reference manager software. After full text review, eligibility of the studies was re-evaluated, excluding the non-eligible studies. Data of studies that were considered eligible after full text review were extracted.
2.4. Data Collection
Data were extracted in a custom-made table for each study. The table includes a section of the basic information of the study design, settings, and funding; then a section for the inclusion and exclusion criteria of the participants; followed by a section for the intervention(s) used and the follow up period; then details about the sampling technique of plaque samples and the analysis platform.
2.5. Risk of Bias in Individual Studies
After data was extracted, the risk of bias of the included studies was assessed. As our review includes different study designs, each type was assessed using its specific tool of risk of bias assessment.
For RCTs, the Cochrane risk of bias assessment tool was used. It includes seven domains: random sequence generation (selection bias), allocation concealment (selection bias), blinding of participants and personnel (performance bias), blinding of outcome assessment (detection bias), incomplete outcome data (attrition bias), selective reporting (reporting bias), and other bias.
In each domain, the risk of bias was judged as “low”, “high”, or “unclear”. A study was considered of “low risk of bias” if all its domains have a low risk of bias; while considered as having an “unclear risk of bias” if one or more of the domains have an unclear risk of bias. “High risk of bias” study has one or more of the domains with a high risk of bias [7].
ROBINS-I tool was used for the assessment of the risk of bias of non-RCTs. It includes seven domains: bias due to confounding, bias in the selection of participants, bias in the classification of interventions, bias due to deviations from intended interventions, bias due to missing data, bias in the measurement of outcomes, and bias in the selection of the reported results.
In each domain, each question should be answered as Yes, Potentially yes, No, Potentially No, or No information. Accordingly, the risk of bias of the domain will be judged as “low”, “moderate”, “serious”, “critical”, or “no information”. Then the overall risk of bias was judged based on the risk of bias of each domain, assessing the study as having “low”, “moderate”, “serious”, “critical” risk of bias, or “no information” [8].
For before-and-after studies, the quality assessment tool for quantitative studies was used [9]. It consists of six domains, namely selection bias, study design, confounders, blinding, data collection method, withdrawals, and dropouts. Each domain is assessed as being “weak”, “moderate”, or “strong”; and accordingly, the global rating of the study is assessed [10].
All the steps of the review-starting from running the search, screening of the search results by title and abstract then by full text, data extraction, and ending by the risk of bias assessment- were performed by the two review authors independently. Any conflicts in the decisions of the two authors were resolved by discussion.
2.6. Synthesis of Results
The included studies were categorized according to the type of intervention. The studies that tested the same type of intervention were gathered in a table to clarify the effect size of intervention in each study. Meta-analysis was planned to reach the pooled effect size of the interventions but was rather not possible to all the outcomes due to the substantial heterogeneity of the methodology of the included studies. For the outcomes in which meta-analysis was possible, it was performed using RevMan 5.3 software. Mean differences were used and a pooled estimate has been calculated.
3. RESULTS
3.1. Study Selection
After pooling of all the searched results and omitting the duplicates, 2700 records were screened by title and abstract for eligibility. Forty of these records were considered eligible by title and abstract, among which ten records were eligible by full text (Fig. 1).
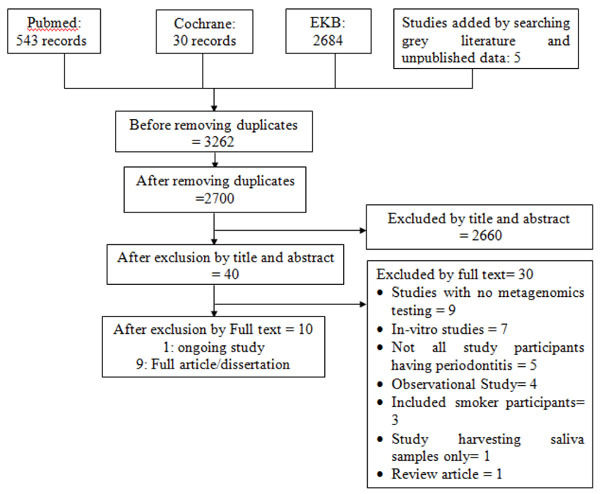
Thirty records were excluded after reviewing the full text: nine of them did not include metagenomics testing as an outcome [11-19], seven were in vitro studies [20-26], five included participants who did not suffer from periodontitis [27-31], four were observational studies [32-35], three included smoker patients [6, 36, 37], one only assessed saliva samples [38], and one was a review article [39]. The 30 records excluded by full text are listed along with the reasons for their exclusion in Appendix A.
The ten included records include 5 RCTs [40-44], 3 before-and-after [45-47], 1 non-RCT studies [48], and 1 protocol of an ongoing study [49]. The detailed data of the included studies are listed in Appendix B.
3.2. Study Characteristics
Among the 9 included studies, 2 publications [44, 45] were found to be pilot studies or preliminary results of 2 fully implemented studies [41, 46]. In 2012, the dissertation of Chang [45] included only 4 patients who received non-surgical periodontal treatment. Chang assessed the subgingival microbiological community after non-surgical periodontal treatment using both Sanger sequencing and 16S rRNA metagenomics using Shotgun sequencing. The publication of Shi et al. [46] included 12 patients, followed the same methodological steps and reported the same outcomes.
Also in 2102, Jünemann et al. [44] performed a pilot study including only 4 participants; while in 2018, the final publication by Hagenfeld et al. [41] included 96 patients with major changes in the microbiological analysis methods. They compared the effect of conventional non-surgical periodontal treatment alone to that combined with antibiotic coverage.
Similarly, a single before-and-after study [47] assessed the effect of conventional non-surgical periodontal treatment as well.
Other than the 5 aforementioned included studies, each of the 4 remaining studies assessed a different intervention, namely anti-adhesive and anti-bacterial toothpastes [40], lactoferrin and lactoperoxidase probiotics [43], and sodium hypochlorite mouth rinse [48]. Lastly, Queiroz et al. [42] included patients with furcation involvement as a specific cohort of chronic periodontitis. They assessed the effect of bone, enamel matrix derivatives compared to both combined together in the treatment of cases of furcation involvement.
3.3. Risk of Bias Assessment
The filled forms of the risk of bias assessment tools of the 9 included studies are listed in Appendix C and summarized in Fig. (2).
As the study of Hagenfeld et al. in 2019 [40] was a microbiological analysis of the samples of the previously published RCT of Harks et al. [50], the assessment of the risk of bias was based on both studies.
Regarding the RCTs, except for the study of Hagenfeld et al. [40, 50], all the studies did not state the method of random sequence generation; instead, they just stated that the study was randomized. The allocation concealment was only clarified by Hagenfeld et al. [40, 50] and Queiroz et al. [42]. Similarly, the blinding of participants and personnel was not explained except in the study of Nakano et al. [43] and was not applicable in the study of Queiroz et al. [42]. On the other hand, the outcome assessor blinding was not stated in any of the 5 studies.
Lastly, the incomplete outcome data was considered of high risk of bias in 2 studies [40, 43]. In the case of Hagenfeld et al. [40], the primary study by Harks et al. [50] recruited 70 patients while the analysis performed by Hagenfeld et al. [40] only included the samples of 41 patients with no justification of the change in the number of samples. Whereas Nakano et al. [43] included patients who were edentulous from whom plaque sampling was not applicable, besides the participants who violated the exclusion criteria were excluded from the analysis rather than including their data in an intention-to-treat analysis.
The solitary non-RCT included [48] had low risk of bias in all domains except for serious risk of bias in classification of interventions; as no criteria were stated according to which each participant was allocated in either intervention or in control groups.
Shifting to the included before-and-after studies [45-47], none of the 3 studies stated the number of patients who agreed versus those who did not agree to participate in the study. Therefore, selection bias was considered of high risk. Regarding the blinding domain, the tool would suggest the studies to be of high risk of bias; however, the reviewers decided that the blinding would not affect the performance of the participants or the personnel or the outcome values. Therefore, the blinding domain was assessed for the 3 studies as of low risk.
On the contrary, in one before and after study [46], the tool suggests the study would be of moderate risk of bias; but the reviewers decided to increase the ranking of the risk of bias from 'moderate' to 'high'. The authors of the study only included in the analysis samples of periodontal pockets that responded to treatment. In addition, they did not report the clinical outcome; therefore the study was considered to have a high risk of reporting bias. Furthermore, the intrinsic limitation of the study design of the 3 studies rendered the study design domain of moderate risk of bias.
Other than the above-stated risks of bias in the 9 included studies, all the domains showed low risk of bias. Finally the included studies range between having high [40, 43, 46], serious [48], moderate [45, 47], and unclear [41, 42, 44] risks of bias. None of the included studies proved low risk of bias.
3.4. Effects of Interventions
Among the included studies, 5 questioned the effect of non-surgical periodontal treatment. The 5 studies include an RCT [41] and its pilot RCT study [44], a before and after preliminary results of a dissertation [45] and its full published article [46], and a before and after study [47].

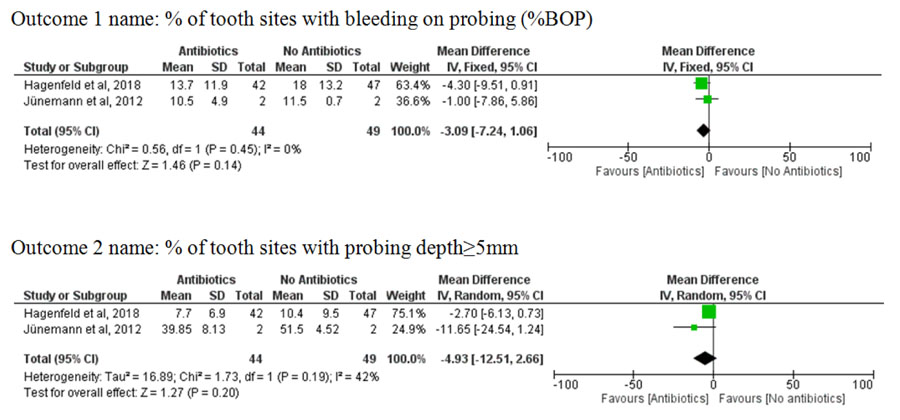
Regarding the RCTs, they assessed the efficacy of non-surgical periodontal treatment compared to it combined with antibiotic coverage. Ion Torrent platform was used in the pilot study of Jünemann et al. [44], yielding limited read lengths and relatively high sequencing errors. Therefore, the study of Hagenfeld et al. [41] shifted to the use of Illumina MiSeq. This methodological difference would not provide reliable effect size if the microbiological results were pooled statistically. Therefore, meta-analysis for the outcomes of microbiological findings was not possible. However, the clinical results were pooled together resulting in a non-significant difference in probing depth (pooled mean difference of %PPD 5 mm = -4.93, 95% CI = -12.51, 2.66) and bleeding on probing (pooled mean difference of %BOP = -3.09, 95% CI = -7.24, 1.06) between prescribing and not prescribing antibiotics in conjunction with non-surgical periodontal treatment (Fig. 3).
In Table 1, the characteristics and effect sizes of each study are reported separately; also, in each RCT, the control group results were calculated and reported assuming it was performed in a before and after study.
On the other hand, the before and after studies did not provide sufficient numerical data for meta-analysis; besides, the study of Yamanaka et al. [47] assessed supragingival samples using pyrosequencing rather than subgingival samples using Shotgun analysis utilized in the other 2 studies [45, 46].
Each of the remaining 4 studies [40, 42, 43, 48] tested a different intervention as shown in Table 1. However, the 4 studies did not provide enough numerical data of the results; they rather preferred its presentation in diagrammatic forms that were not possibly used in the analysis, reviewing the statistical methods and reaching a sound conclusion.
The descriptive microbiological results of the included studies were gathered in Table 2; in which the bacterial community is reviewed in each of the 3 niches discussed in the review: supragingival plaque, subgingival plaque, and subgingival plaque of furcation defects.
| Study | Study Design | No. of Participants | Intervention/s | Follow up Period | Type of Sample & (Plat-Form) | Outcomes (Effect size) | Interpretation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical | Microbiological | ||||||||||
| I. The effect of non-surgical periodontal treatment | |||||||||||
| (Jünemann et al., 2012) [44] | RCT | 4 | I*: Mechanical debridement + 500 mg Amoxicillin + 400 mg Metronidazole VS C*: Mechanical debridement + placebo |
2 months | Subgingival (Ion Torrent) |
Mean diff. %PPD>7mm*= -0.8 %BOP*=-1 %PI*=4.5 Control (before and after) %PPD*=-18.7 %BOP*= -55 %PI*=-46 |
Mean diff. ACE= -58.5 Shannon=-0.48 Simpson=0.025 Evenness= -0.04 Control ACE=28.5 Shannon=0.29 Simpson= -0.03 Evenness=0.035 |
Clinical and microbiological results favor I* C* alone improves clinical but not microbiological results |
|||
| (Hagenfeld et al., 2018) [41] | RCT | 96 | I*: Mechanical debridement + 500 mg Amoxicillin + 400 mg Metronidazole VS Mechanical debridement. + placebo |
2 months | Subgingival (Illumina MiSeq) |
Mean diff. %PPD5mm*= -2.7 %BOP= -4.3 %RAL1.3mm*= 0.1 Control %PPD5= 7.5 %BOP=16.4 |
Mean diff. Richness= -14.91 Evenness= 0 Diversity= -0.1 Dissimilarity= 0.04 Contol Richness= -4.41 Evenness= 0 Diversity= 0.02 Dissimilarity= -0.01 |
Clinical and microbiological results favor I* C* alone improves microbiological but not clinical results |
|||
| (Chang, 2012) [45] | Before and after | 4 | Scaling and root planning + oral hygiene instruct. | Baseline, 4-6 weeks | Subgingival (Shotgun: Illumina GAIIx) |
Mean difference GI*=-1.3 PD*= -2 mm GR*= 1.3mm CAL*= -1.5 mm |
Descriptive data only | I* improves the clinical results and causes a shift in the subgingival microbial community | |||
| (Shi et al., 2015) [46] | Before and after | 12 | Scaling and root planning + oral hygiene instruct. | Baseline, week 4 and 19 | Subgingival (Shotgun: Illumina GAIIx) | Not mentioned; The study only included the clinically resolved cases |
No numbers reported (diversity in resolved< diseased) | According to descriptive data, I* improves the microbiological results | |||
| (Yamanaka et al., 2012) [47] | Before and after | 19 | Scaling and root planning + oral hygiene instruct. | Baseline and 2 years | Saliva and supragingival (pyrosequencing) |
Mean diff. between % of pockets>7mm= -6.98% |
Mean diff. Chao I= -211 ACE=-214 Shannon=- 0.3 |
Intervention improves clinical and microbiological results | |||
| II. The effect of other interventions (Miscellaneous) | |||||||||||
| (Califf et al., 2017) [48] | Non-RCT | 34 | I*: 0.25% sodium hypochlorite rinse VS C*: 15 ml of water rinse |
Baseline, day 14, month 3 | Supra and subgingival plaque (Illumina MiSeq) | Not stated to be an outcome for the study | No numbers reported. | According to descriptive data, I* causes a shift in the plaque microbial community | |||
| (Hagenfeld et al., 2019) [40] | RCT | 41 | I*: zinc-substituted carbonated hydroxyapatite dentifrice VS C*: dentifrice containing an amine fluoride/stannous fluoride |
Baseline, week 4 and 12 | Supra and subgingival plaque (Illumina MiSeq) |
Mean diff PFR*=2.3% PCR*=2.6% GI*=-0.1 BOP*=0.9% PPD*=-0.1 mm GR*=0.2 mm AL*=0.1mm |
No noticeable difference between the 2 groups | C* provides better clinical results than I* and both show similar microbiological effects. | |||
| (Nakano et al., 2017) [43] | RCT | 46 | tablets contain 80 mg of LF+LPO powder (Lactoferrin + lactoperoxidase) VS placebo | Baseline, week 4 and week 8 | Tongue coating and supragingival plaque (Illumina MiSeq) |
No postoperative numbers reported | No postoperative numbers reported (lower diversity in I* than placebo) | I* causes shift in the plaque microbial community lowering the diversity | |||
| (Queiroz et al., 2017) [42] | RCT | 41 with furcation involvement | Bone Vs EMD* VS Bone + EMD | Baseline, 3 and 6 months | Subgingival plaque from the furcation defect (pyrosequencing) |
No data about intervention or control | No numbers reported | - | |||
| Genera that Decreased after Treatment | Type of Treatment | Genera that Increased after Treatment | Type of Treatment |
|---|---|---|---|
| Subgingival Plaque | |||
|
Porphyromonas Tannerella |
NSPT* [44-46] NSPT*+ Antibiotics [41, 44] |
Streptococcus | NSPT* [44-46] NSPT*+ Antibiotics [41, 44] Sodium hypochlorite mouth rinse [48] |
| Filifactor Treponema | NSPT* [45, 46] NSPT*+ Antibiotics [41, 44] |
Prevotella Selenomonas |
NSPT* [44, 45] NSPT*+ Antibiotics [44] Sodium hypochlorite mouth rinse [48] |
| Rothia Selenomonas | NSPT* [44] NSPT*+ Antibiotics [41, 44] |
Actinomyces Capnocytophaga | NSPT* (15-17) NSPT*+ Antibiotics [44] |
| Kingella | NSPT* [41] NSPT*+ Antibiotics [41] |
Neisseria Rothia |
NSPT* (15,17) NSPT*+ Antibiotics [44] |
| Eubacterium | NSPT* [44] NSPT*+ Antibiotics [44] |
Veillonella | NSPT* [45, 46] NSPT*+ Antibiotics [41] |
|
Capnocytophaga Eikenella Fretibacterium Oceanivirga |
NSPT*+ Antibiotics [41] | Corynebacterium | NSPT* [45, 46] NSPT*+ Antibiotics [44] |
|
Gemella Leptotrichia Neisseria |
NSPT* [45] | Campylobacter Fusobacterium | NSPT* [45] |
| Dialister | NSPT* [45, 46] | Cardiobacterium | NSPT* [46] |
| - | - | Derxia | NSPT*+ Antibiotics [44] |
| - | - |
Aerococcus Dialister Slackia |
Sodium hypochlorite mouth rinse [48] |
| Supragingival plaque | |||
|
Fusobacterium Kingella Neisseria Porphyromonas |
NSPT* [47] |
Actinomyces Corynebacterium Rothia |
NSPT* [47] |
| Capnocytophaga | NSPT* [47] Probiotic tablets [43] |
- | - |
|
Camylobacter Enterobacter Leptotrichia Prevotella |
Probiotic tablets [43] |
Bifidobacterium Clostridium Streptococcus |
Probiotic tablets [43] |
| In Furcation defects (Subgingival) | |||
|
Filifactor Fusobacterium Streptococcus Veillonella |
EMD* + Bone [42] |
Camylobacter Parvimonas Pseudomonas Selenomonas |
EMD* + Bone [42] |
| Actinomyces | EMD* [42] | - | - |
4. DISCUSSION
Oral microbiota plays a major role in the pathogenesis of chronic periodontitis [6]. Therefore, we performed this systematic review aiming to assess the effect of periodontal treatments on oral microbiota. Based on the eligibility criteria of the review, we included 9 studies that performed metagenomic analysis after testing certain interventions for the treatment of chronic periodontitis.
The included studies addressed the effect of non-surgical periodontal treatment [45-47], antibiotics in conjunction with non-surgical periodontal treatment [41, 44], different toothpaste ingredients [40], sodium hypochlorite rinse [48], and probiotic tablets [43]. Lastly, a single included study [42] addressed the microbiome of subgingival plaque in cases of furcation involvement after grafting with bone versus enamel matrix derivative versus bone together with enamel matrix derivative.
The review concluded that antibiotic use in addition to non-surgical periodontal treatment insignificantly improves the clinical and microbiological outcomes in comparison to non-surgical periodontal treatment. The use of toothpaste containing amine fluoride/stannous fluoride also proved positive clinical and microbiological results.
Substantial heterogeneity was detected in the included studies regarding study design, type of treatment, sampling technique, sample type, length of the follow-up period, method of statistical analysis, and methods of data reporting. Considering all these aspects of heterogeneity, meta-analysis for all the outcomes of all the included studies was not applicable. Therefore, a meta-analysis was performed only when clinical and methodological heterogeneity allowed for a meaningful pooling of the results; that was for the clinical outcomes of two of the included studies. Otherwise, most of the review results are discussed qualitatively.
The results proved clinical improvement of chronic periodontitis in response to non-surgical periodontal treatment [44-47], antibiotics in conjunction with non-surgical periodontal treatment [41, 44], and toothpaste containing amine fluoride/stannous fluoride [40]. However, the clinical response was found statistically insignificant when comparing non-surgical periodontal treatment with and without antibiotic coverage. The difference in probing depth (pooled mean difference of %PPD 5 mm= -4.93, 95% CI = -12.51, 2.66) and bleeding on probing (pooled mean difference of %BOP = -3.09, 95% CI = -7.24, 1.06) showed insignificant results.
Regarding the microbiological results and based on the scarce numbers reported by the included studies, the diversity of bacterial species was reported to be lowered by non-surgical periodontal treatment [47] and antibiotics in conjunction with non-surgical periodontal treatment [41].
Therefore, the available evidence suggests antibiotic use in conjunction with non-surgical periodontal treatment does not add a significant benefit in the clinical or the microbiological outcomes. Consequently, periodontists are advised to judge the benefits, which are insignificant, versus the risks, including antibiotic resistance, when prescribing antibiotics in cases of chronic periodontitis.
Subgingival plaque community has shown a decrease in periodontitis-associated genera [3] (Porphyromonas, Tannerella, Treponema, and Filifactor) in response to non-surgical periodontal treatment with and without antibiotic therapy. Besides, other genera showed an increase in abundance after treatment (Streptococcus, Prevotella, Selenomonas, Actinomyces Capnocytophaga, Neisseria Rothia, Veillonella, and Corynebacterium). This may be explained by the intermingled relationships between bacteria ranging between growth dependency, synergy, antagonism, and mutual reliance for growth and survival [4]. Therefore, as some species and genera are associated with the disease state, others may be linked to the resolved state.
After effective periodontal treatment, a diseased periodontal pocket passes through a lengthy process of bacterial community shift from the diseased to the resolved rather than the naïve state. Meanwhile, clinical improvement can be noticed that is not coinciding with microbiological balance yet [45].
Accordingly, microbiological results of the included studies were not fully coinciding together as the follow-up period differs among them. It ranged between 6 weeks [45], 2 months [41, 43, 44], 3 months [48], 6 months [40, 42], and 2 years [46, 47]. Therefore, the state of resolution of the microbiome was not always tested at the same time points; producing heterogeneous results.
Likewise, the supragingival plaque community showed a decrease in Porphyromonas, Kingella, and Capnocytophaga; and an increase in Actinomyces, Corynebacterium, and Rothia after treatment.
On the other hand, the community of the furcation defect showed a different shift after surgical treatment. Filifactor Fusobacterium, Streptococcus, and Veillonella were reduced in number; while Camylobacte, Parvimona, Pseudomonas, and Selenomonas show abundance.
Being 3 different niches, the difference in their normal and pathological inhabitants is a normal finding. However, due to the scarcity of data in the literature regarding the metagenomic assessment of periodontal treatments, no solid conclusion could be reached so far to prove the perfect community for each of the above-mentioned niches and the method to maintain it.
Other systematic reviews have been performed, concerning metagenomics in periodontal disease. A valuable systematic review [3] investigated the microbiota composition in periodontal disease. They included 11 case-control studies that compared plaque samples from patients having periodontitis to normal controls. The results of the review ensured the association of some bacterial species to periodontitis; namely, Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. The results also detected 3 newly identified genera that could be considered as periodontal pathogens: Desulfobulbus spp., Filifactor alocis, and TM7 spp [3].
The review differs from ours in its scope, aim, and types of studies included; however, it paved the path for our review by clarifying the bacterial communities in supragingival and subgingival plaque. Therefore, their work made it possible for our review to assess the effectiveness of various interventions to restore the normal microbiological balance.
Another systematic review questioned the effect of tobacco smoking on periodontal microorganisms [51]. The review concluded an association between smoking and subgingival dysbiosis. It also highlighted that this dysbiosis impairs periodontal healing, and therefore emphasized the benefits of treatment of tobacco dependence in order to obtain a healthy subgingival ecosystem that responds to treatment. The aforementioned review included all types of studies that used any type of genetic analysis of the oral microbiome in smoker patients having periodontitis. As our review included only interventional studies recruiting non-smoker patients, the results of the 2 systematic reviews cannot be compared.
Our review faced some limitations; including the small number of the available eligible studies, and the risk of bias of the included studies which questions the level of evidence of the results. None of the included studies proved low risk of bias, therefore the risk of bias should be considered when viewing the final results. Consequently, we would recommend further clinical studies to properly plan and report all the study details based on the reporting guidelines to improve the level of the available evidence. Another limitation of the review was the substantial heterogeneity of the included studies preventing the gathering of all available data. Therefore, a standardized protocol is recommended for future clinical trials.
CONCLUSION
Based on the very limited number of eligible studies included in the review, together with their substantial heterogeneity, the level of the available evidence is not sufficient to draw a solid conclusion about the effect of periodontal treatments on the periodontal microbiome. Further well-planned future studies with standardized protocols are needed to get a clear conclusive answer to the review question.
AUTHORS' CONTRIBUTIONS
Both authors contributed equally in all the steps of the protocol: Drafting the manuscript of the protocol, developing the selection criteria, data extraction criteria, search strategy, and approving the final version of the protocol.
CONSENT FOR PUBLICATION
Not applicable.
STANDARD OF REPORTING
This study follows the protocol established by PRISMA guideline.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Dr. Fat'heya Zahran, (Professor of Oral Medicine and Periodontology, Cairo University) for her outstanding help and generous advice.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Website along with the published article.


