All published articles of this journal are available on ScienceDirect.
Development of In Vitro Denture Biofilm Models for Halitosis Related Bacteria and their Application in Testing the Efficacy of Antimicrobial Agents
Abstract
Objective : Since dentures can serve as a reservoir for halitosis-causing oral bacteria, halitosis development is a concern for denture wearers. In this study, we surveyed the prevalence of four selected halitosis-related species (Fusobacterium nucleatum, Tannerella forsythia, Veillonella atypica and Klebsiella pneumoniae) in clinical denture plaque samples, and developed denture biofilm models for these species in vitro to facilitate assessment of antimicrobial treatment efficacy. Design : Denture plaque from ten healthy and ten denture stomatitis patients was screened for the presence of aforementioned four species by PCR. Biofilm formation by these halitosis-associated species on the surfaces of denture base resin (DBR) discs was evaluated by crystal violet staining and confocal laser scanning microscopy. The efficacy of denture cleanser treatment on these mono-species biofilms was evaluated by colony counting. Results : 80% of the subjects in the denture stomatitis group and 60% in the healthy group contained at least one of the targeted halitosis-related species in their denture plaque. All halitosis species tested were able to form biofilms on DBR disc surfaces to varying degrees. These in vitro mono-species resin biofilm models were used to evaluate the efficacy of denture cleansers, which exhibited differential efficacies. When forming biofilms on resin surfaces, the halitosis-related species displayed enhanced resistance to denture cleansers compared with their planktonic counterparts. Conclusion : The four selected halitosis-related bacterial species examined in this study are present on the majority of dentures. The mono-species biofilm models established on DBR discs for these species are an efficient screening tool for dental product evaluation.
1. INTRODUCTION
Halitosis, also known as oral malodor or bad breath, is foul-smelling breath exhaled from the oral cavity. It affects about one-third of the population and has profound physical, social and psychological impacts on affected individuals [1]. Halitosis is mainly due to the metabolic products of oral bacteria using proteinaceous compounds from saliva, gingival crevicular fluid, and epithelial cells as substrates [1, 2]. Volatile sulfur compounds (VSCs), especially hydrogen sulfide (H2S), methyl mercaptan (CH3SH), and dimethyl sulfide [(CH3)2S] are the major molecules implicated in oral halitosis [3]. In addition to these VSCs, other malodorous molecules including short-chain fatty acids as well as polyamines produced by anaerobic microbes residing on the surface of the tongue and in the periodontal pockets, contribute to halitosis[2].
A variety of oral bacteria have been shown to produce malodorous compounds [3-8]. Tannerella forsythia (formerly Bacteroides forsythus), Fusobacterium nucleatum, Klebsiella pneumoniae, Veillonella atypica, Porphyromonas gingivalis, Prevotella intermedia, Treponema denticola, Atopobium parvulum and Eubacterium sulci are common species frequently isolated from halitosis patients [3-8]. These malodorous compound-producing bacteria are present at various sites in the oral cavity, particularly on the dorsum of the tongue, saliva, periodontal pockets, dentures, and dental restoration sites.
Recent studies found a strong association between the degree of denture hygiene and halitosis, with higher levels of VSCs detected among denture-wearing subjects, and particularly in elderly individuals wearing dentures overnight and subjects with denture stomatitis [4, 9-11]. Taken together, these findings suggested that halitosis-related oral bacteria might be able to colonize denture surfaces, become residents of the denture plaque microbial communities and thus contribute to denture-associated halitosis. However, very few studies have investigated the colonization and biofilm formation of halitosis-related oral species on denture surfaces. Meanwhile, with the increased demand for dental hygiene products, simple and reproducible denture biofilm models of halitosis-associated oral species are needed to test the effectiveness of these products, particularly denture cleansers, against denture-associated odor-causing bacteria.
In this study, we evaluated the prevalence of the four selected halitosis-related bacterial species in clinical denture plaque samples of denture stomatitis patients and healthy subjects. Furthermore, mono-species denture base resin (DBR) biofilm models were developed for these species and used for evaluating the efficacy of denture cleansers.
2. MATERIALS AND METHODS
2.1. Subjects and Sampling
Two groups of subjects were included in this study and informed consent was obtained prior to the collection of samples. Group 1 contained ten healthy denture wearers (five women and five men; mean age 69.8±4.7 years); group 2 included ten denture wearers diagnosed with denture stomatitis (five women and five men; mean age 61.6±12.0 years) based on the guideline for denture stomatitis diagnosis [12]. These individuals had not used antibiotics for at least 3 months before sample collection. Subjects had not been treated for any systemic disease nor were they taking any prescription or nonprescription medications.
Subjects were asked to wear dentures for at least 3 hrs before samples were taken. For sample collection, sterile toothpicks (Fisher, USA) were used to scrape the plaque from the surface of the denture that was in contact with the oral mucosa. Collected plaque samples were placed into separate microcentrifuge tubes containing 0.5 ml of 80% ethanol and stored at -20°C until DNA extraction. The QIAamp DNA Micro Kit (Qiagen, USA) was used to extract DNA from collected plaque samples according to manufacturer’s instructions. DNA quality and quantity were checked with a NanoDrop 2000 spectrophotometer (Thermo, USA).
2.2. Detection of Halitosis-related Species by Polymerase Chain Reaction (PCR)
PCR was employed to detect the presence of the four selected halitosis-related bacterial species K. pneumoniae, V. atypica, T. forsythia and F. nucleatum within the plaque samples. Bacterial species-specific primers were designed based on previous studies[13-15].
For the PCR reactions, 10 ng DNA isolated from plaque samples was used as template in a 25 μl reaction mixture containing 1x PCR buffer, 1.5mM MgCl2, 0.1mM each of the four dNTPs, 1 unit Taq DNA polymerase (Invitrogen, USA) and 1 μM of each primer. The PCR reaction conditions with optimized cycle and annealing temperature for each specific primer sets were as follows: for K. pneumoniae, specific primers Pf (5′-ATT TGA AGA GGT TGC AAA CGA T-3′) and Pr2 (5′-CCG AAG ATG TTT CAC TTC TGA TT-3′) were used; the reaction conditions were 94°C for 10 min; 35 cycles of 94°C for 30 s, 57°C for 20 s, and 72°C for 20 sec, and a final extension at 72°C for 10 min. For V. atypica, specific primer set ATYF (5′- TCT CTT GTT GAA GAA TTA GAA CGC-3′) and VR (5′- GTGTAACAAGGGAGTACGGACC-3′) was used; the PCR conditions were 94°C for 15 min, 20 cycles of 92°C for 1 min, 57°C for 1 min, and 72°C for 1 min, followed by 5 min at 72°C. For T. forsythia, specific primers 5′- GCGTATGTAACCTGCCCGCA-3′ and 5′- TGCTTCAGTGTCAGTTATACCT-3′ were used, and the reaction conditions were 94°C for 3 min, 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min, followed by 72°C for 5 min. For F. nucleatum, specific primer set 5′- AGAGTTTGATCCTGGCTCAG-3′ and 5′-GTCATCGTG CACACAGAATTGCTG -3’ was used; the PCR conditions were 94 °C for 3 min; 23 cycles of 94°C for 1 min, 61.5°C for 1 min, and 72°C for 30 sec, and a final extension at 72°C for 5 min.
PCR products were analyzed by agarose gel electrophoresis (1% w/v) in 1×TAE buffer (40 mM Tris–HCl, 1.18 ml acetic acid, 2 mM EDTA, pH 8.0) and a constant voltage of 100V was applied. Images were digitally recorded using the Molecular Imager Gel Documentation system (Bio-Rad, USA) to evaluate the presence of the amplified DNA product.
2.3. Strains and Growth Conditions
K. pneumoniae IA 565 is a clinical isolate obtained from the University of Iowa Hospitals and Clinics Special Microbiology Laboratory. The strain was grown in LB medium (Tryptone 10g/L Yeast Extract 5g/L NaCl 10g/L) and incubated at 37°C in an aerobic incubator on a rotary shaker operating at 200 rpm. T. forsythia ATCC 43037, V. atypica PK 1910 and F. nucleatum ATCC 23726 were grown anaerobically overnight at 37°C in TF medium, TH medium containing 0.06% (w/v) lactic acid and Columbia broth (Difco, USA), respectively [16].
2.4. DBR Discs
The acrylic DBR discs used in this study were manufactured as previously described[17]. All resin discs were disinfected with 2% (v/v) sodium hypochlorite solution (Sigma-Aldrich, USA) for 10 min and immersed in sterilized ddH2O overnight prior to microbial inoculation.
2.5. Growth of Biofilms on DBR Disc Surfaces
Exponentially growing cultures of the four halitosis-associated bacterial species were diluted in their respective culture medium to a final OD600 of 0.1 Two ml of the respective diluted cultures of the different species were added to each well of 12-well plates (Thermo, USA) containing a DBR disc with the rough surfaces exposed. Wells with growth medium alone served as controls. Plates containing cultures of T. forsythia, F. nucleatum, or V. atypica were incubated at 37°C anaerobically for 24 hrs, while cultures of K. pneumoniae were grown aerobically at 37°C for 24 hrs.
2.6. Crystal Violet Staining of Biofilms on DBR Disc Surfaces
Biofilms formed on the DBR discs were stained with crystal violet as described previously [18]. Briefly, DBR discs were washed twice with phosphate-buffered saline (PBS), air dried for 30 min, and stained with 0.4% (w/v) crystal violet in distilled water for 20 min. The stained discs were gently washed three times with sterilized distilled water and air dried for 30 min before being photographed. Images were taken with a D50 digital camera (Nikon, Japan).
2.7. Observation of Biofilms with Confocal Laser Scanning Microscopy (CLSM)
Different methods have been used to study biofilm formation including CLSM, AFM [19]. In our study, CLSM has been employed to reveal the biofilm structure as well as the cell viability within biofilms. Biofilms grown on the rough surface of the DBR discs were treated with Polident® experimental formula M138-12 (GlaxoSmithKline, UK) or Efferdent® antibacterial denture cleanser (Prestige brands, USA) for 5 min according to manufacturer’s recommendations, and untreated controls were stained with 10 μM of the cell-permeable fluorescent stain SYTO 59 (labeling of all cells) and 10 μM of the cell-impermeable fluorescent stain SYTOX Green (labeling of cells with compromised cell walls only) (Invitrogen, USA) in PBS buffer for 20 min at room temperature in the dark before visualization [17]. All biofilm images were collected with a Zeiss LSM 5 PASCAL CLSM using LSM 5 PASCAL software (Zeiss, Germany). Excitation at 633 nm with an argon laser in combination with a 650 nm band-pass emission filter was used for SYTO 59 fluorescence imaging. SYTOX Green signals were visualized using 488 nm excitation with a helium-neon laser and a 503-530 nm band-pass emission filter.
2.8. Evaluation of the Treatment Efficacy of Denture Cleansers in Planktonic and Biofilm Conditions
Biofilms grown on DBR discs were washed three times with PBS buffer before treatment. Two different Polident® denture cleansing tablets (commercialized formulation 50767 and the experimental formulation M138-12) (GlaxoSmithKline, UK) and Efferdent® antibacterial denture cleanser (Prestige Brands, USA) were separately dissolved in 150 ml 40°C distilled water according to manufacturer’s instructions. Each DBR disc with attached biofilms was immersed in 50 ml of the treatment solution for 5 min for all three denture cleansers and an additional experiment with a 15 min immersion period for Efferdent® antibacterial denture cleanser was performed to follow manufacturer’s recommendations. DBR discs with attached biofilms treated with PBS buffer served as negative control. After treatment, the DBR discs were washed three times with PBS buffer and subjected to mechanical disruption of the attached biofilms, serial dilution and plating as previously described to obtain colony forming unit counts[17].
One ml of exponentially growing planktonic cultures (OD600 of 1) was washed three times with PBS buffer and pelleted by centrifugation at 16,000 xg for 2 min. One tablet each of the three different denture cleansers tested was predissolved in 150 ml 40°C water for 2 min and 1 ml of the treatment solution was used to resuspend the pelleted cells. After 5 min or 15 min (for the additional treatment with Efferdent®) of treatment (3 or 13 min incubation with additional 2 min centrifugation at 16,000 xg), cells were washed three times with PBS buffer prior to being subjected to serial dilution and plating.
Treatment efficacy was evaluated by comparing the surviving colony-forming units (CFU) on LB agar plates (for K. pneumoniae), Columbia agar plates supplemented with 5% sheep blood (Colorado Serum company, USA) (for F. nucleatum), TH agar plates supplemented with 0.06% lactic acid (for V. atypica), and TF agar plates supplemented with 5% sheep blood (for T. forsythia) with PBS-treated control group.
3. RESULTS
3.1. The Prevalence of Four Halitosis-related Strains in Denture Plaque
The aforementioned four halitosis-related bacterial species were chosen as targets for PCR screening among 20 clinical denture plaque samples (10 each for healthy and stomatitis subjects). Among the four targeted species, K. pneumoniae was only detected in two of the ten subjects in the stomatitis group, while the remaining three species were present in both groups (Table 1). Their prevalence in healthy and denture stomatitis plaque was 60% and 40% for F. nucleatum, 30% and 40% for T. forsythia, 30% and 10% for V. atypica, 0% and 20% for K. pneumoniae; respectively (Table 1). Furthermore, 80% of the subjects in the denture stomatitis group and 60% in the healthy group contained at least one of the targeted halitosis-related species in their denture plaque (Table 1). Due to small sample sizes, Fisher-exact chi-square test was used to examine the difference between stomatitis and healthy groups across the species-specific primers (p=0.39).
3.2. Biofilm Formation of Halitosis-related Oral Bacterial Strains on DBR Discs with Rough Surfaces
Crystal violet staining revealed that under the conditions described in Materials and Methods, all four species were capable of forming biofilms on these DBR disc surfaces, although to different extents, as manifested by the different intensities of purple staining (Fig. 1). The biofilm architecture of mono-species biofilms and live/dead distribution of the bacteria within the biofilms was further investigated by CLSM.
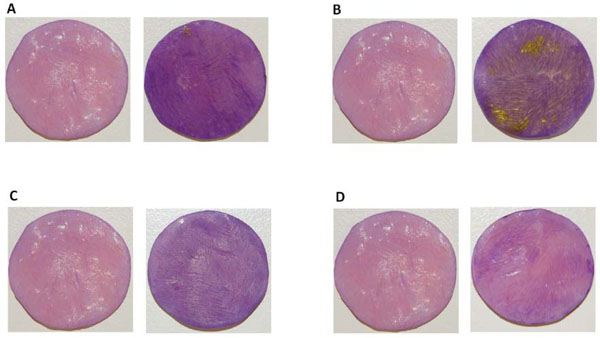
Biofilm formation on DBR disc surfaces. Representative images after crystal violet staining of biofilms formed on DBR discs by F. nucleatum ATCC 23726 (A) T. forsythia ATCC 43037 (B) V. atypica PK 1910 (C) K. pneumoniae IA 565 (D) For each set, the image on the left was a control DBR disc without biofilms growing on the surface, the image on the right was a DBR disc with biofilms growing on the surface. The experiment was performed in triplicate.
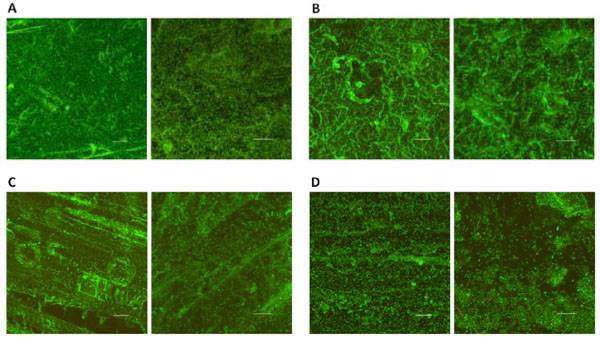
CLSM images of biofilms on DBR disc surfaces. Biofilm formation of F. nucleatum ATCC 23726 (A) T. forsythia ATCC 43037 (B) V. atypica PK 1910 (C) K. pneumoniae IA 565 (D) on DBR discs. For each set, the image on the left was taken through a 20x objective (Scale bar, 50 μm); while the image on the right was taken through a 63x objective (Scale bar, 20 μm). Four random fields of view were examined for each sample and representative images are shown.
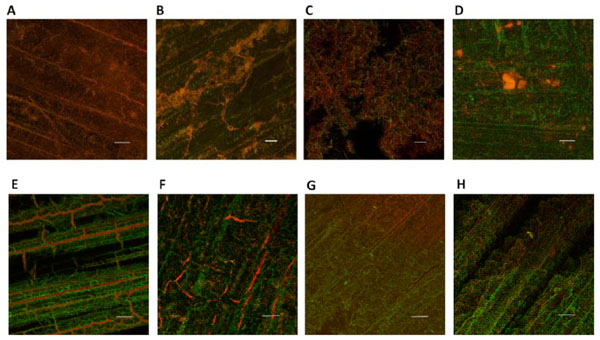
CLSM images of F. nucleatum (A, E) T. forsythia (B, F) V. atypica (C, G) K. pneumoniae (D, H) biofilms formed on DBR disc surfaces after treatment with Polident® experimental formulation M138-12 (upper panel) and Efferdent® (lower panel), respectively. Denture biofilms were stained with SYTO59 and SYTOX Green as described in Materials and Methods, and examined by CLSM to reveal the live (green) and dead (red) cell population following treatment. Four random fields of view were examined for each sample and representative images taken through a 20x objective are shown (Scale bar, 50 μm).
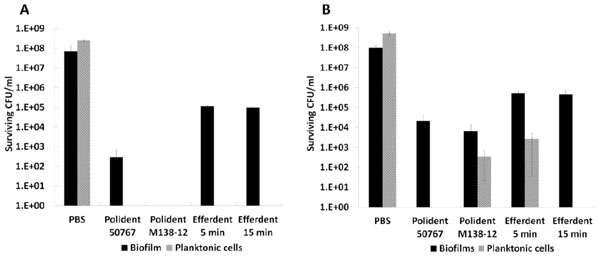
Antimicrobial treatment efficacy against biofilms formed on DBR disc surfaces and planktonic cells. The biofilms and planktonic cells of F. nucleatum (A) and K. pneumoniae (B) were treated with denture cleansers as described in the Materials and Methods. The viability of biofilms after treatment was assessed by colony-forming units (CFU) on agar plates. Standard error of three replicates is presented.
CLSM analyses disclosed distinct biofilm architectures for each of the halitosis-related strains tested. F. nucleatum ATCC 23726 formed biofilms with a smooth and homogeneous structure almost completely covering the irregular DBR disc surface (Fig. 2A). A similar surface coverage was observed for T. forsythia ATCC 43037; however, the biofilms formed were less dense compared to F. nucleatum with great variations in overall thickness (Fig. 2B). Surface coverage by V. atypica PK 1910 was incomplete and less compact compared with mono-species biofilms of F. nucleatum and T. forsythia (Fig. 2C). Among the bacterial strains tested K. pneumoniae IA 565 displayed the least biofilm forming ability. Colonization was limited to thin, “patch-like” structures leaving most of the surface uncovered (Fig. 2D). The majority of the bacteria cells within all mono-species biofilms were alive as indicated by predominantly green fluorescent (live cell staining with SYTO59) and very limited red fluorescent (dead cell staining with SYTOX Green) labeling.
3.3. Evaluating the Antimicrobial Efficacy of Denture Cleansers Against DBR Disc Surface-associated Biofilms and Planktonic Cells of Halitosis-related Strains
For a quantitative assessment of treatment efficacy, the DBR disc based biofilm model developed above as well as planktonic cells of K. pneumoniae and F. nucleatum were subjected to treatment as above and CFU counts were used as a read-out for treatment efficacy. F. nucleatum mono-species biofilms grown on DBR discs were most sensitive to the experimental formula Polident® M138-12 treatment with no viable cells being detected after 5 min incubation. Treatment with Polident® 50767 for the same time period resulted in 102-103 surviving CFU, while 105-106 surviving CFU were still recovered after 5 min and 15 min treatment with Efferdent® (Fig. 4A). In contrast, planktonic F. nucleatum cells exhibited similar sensitivities to all three formulas, with no surviving CFU being detected after 5 min treatment as well as the additional 15 min treatment with Efferdent® (Fig. 4A). While Polident® 50767 and the experimental formula Polident® M138-12 demonstrated similar killing efficacy toward K. pneumoniae denture biofilms, resulting in about 4 orders of magnitude reduction (from ~108 to ~104) in cell viability (t-test, P<0.01) after 5 min treatment, Efferdent® manifested about 3 orders of magnitude reduction (from ~108 to ~105) in cell viability (t-test, P<0.01) after 5 min and 15 min treatment (Fig. 4B). The planktonic form of K. pneumoniae was more sensitive than its biofilm counterpart, with no viable cells being detected after a 5 min treatment with Polident® 50767 or a 15 min treatment with Efferdent®. Three min treatments with the experimental formula Polident® M138-12 or Efferdent® resulted in 102-103 and 103-104 surviving CFU, respectively (Fig. 4B). Quantitative values for killing efficacy were not determined for V. atypica and T. forsythia since CFU recovery on the recommended agar medium plates exhibited large variability under our experimental conditions.
Mono-species DBR-attached biofilms formed by halitosis-related bacterial species were used to evaluate the efficacy of denture cleansers. Biofilms were stained with SYTO59 and SYTOX Green and examined by CLSM to reveal the live/dead cell distribution following the different treatments as described in Material and Methods. Compared to PBS-treated mono-species denture biofilms in which the majority of the cells were alive, treatment with the experimental formula Polident® M138-12 almost completely killed the bacterial cells within mono-species biofilms formed on DBR discs by F. nucleatum, T. forsythia and V. atypica (Figs. 3A, 3B and 3C). In contrast, the same treatment of K. pneumoniae biofilms yielded less apparent killing (Fig. 3D). Treatment with Efferdent® did not result in complete killing of bacterial cells within any of the mono-species denture biofilms formed by F. nucleatum, T. forsythia, V. atypica, or K. pneumoniae within 5 min of treatment time (Figs. 3E, 3F, 3G and 3H).
4. DISCUSSION
Denture–related halitosis poses a serious problem, particularly for elderly denture-wearers[10]. The colonization of denture surfaces with odor-producing oral bacteria has been suspected to play an important role in causing denture-associated halitosis [4, 9-11]. However, the ability of halitosis-related bacteria to colonize and form biofilms on denture surfaces remains poorly understood.
In this study, clinical denture plaque samples were screened for the presence of four selected oral bacterial species that have previously been implicated in halitosis and are known to produce malodorous compounds [3-7]. A high percentage of subjects contained at least one of these halitosis-related species within their denture plaque, although no significant difference was observed between the stomatitis and healthy group (Table 1). The high prevalence of these halitosis-related microbes within denture plaques from both healthy (60%) and denture stomatitis patients (80%) suggested that these bacterial species were able to colonize denture surfaces and therefore could have biofilm-forming ability.
Our study further demonstrated that all four tested halitosis-related species were indeed capable of biofilm formation on the surface of discs manufactured from PMMA, the most commonly used denture material (Fig. 1). The ability to colonize and develop biofilms on denture surfaces not only enables these bacteria to exploit an additional growth niche; biofilms also provide an environment that is more protected against the host immune defense as well as other external stresses such as antimicrobial agents [20, 21]. In cases of poor oral hygiene, dentures could serve as a reservoir for these odor-producing bacteria and result in persistent halitosis.
Our result also revealed different biofilm formation abilities and various biofilm structures among the tested species, with F. nucleatum and T. forsythia exhibiting greater biofilm coverage of PMMA surfaces. This is consistent with our previous studies showing that different bacterial species displayed differential colonization of the same surfaces, and surfaces of different physical properties greatly affected the biofilm formation ability of the same bacterial strains [17].
The observed higher resistance of denture biofilm cells to Polident® and Efferdent® antibacterial denture cleansers compared to their planktonic counterparts is not surprising, since biofilm cells are generally more resilient against external stresses including antimicrobial treatments (Fig. 4)[20]. This highlights the importance of testing antimicrobial activities against relevant biofilm cells rather than planktonic microbes. A variety of dental products, including denture cleansers, which often contain antimicrobial compounds, have been developed for treating denture-related halitosis by reducing or eliminating odor-producing bacteria [22]. Our study demonstrated the successful application of mono-species denture biofilm models for halitosis-related bacteria in evaluating the efficacy of such cleansers. Differential killing of mono-species biofilms of different species was observed for the same denture cleanser formula. Meanwhile, the three denture cleansers tested in this studies exhibited different killing efficacies when tested with the same bacterial species. Our findings confirm that these denture biofilm models can serve as a useful model system for screening and evaluation of dental products, particularly products for treating denture-related halitosis. Furthermore, the mono-species DBR biofilm model developed in this study could be expanded to establish a more clinically relevant multispecies halitosis-related biofilm model for dental product evaluation, as well as for pathogenesis studies of denture-related halitosis.
CONCLUSION
Four selected halitosis-related bacterial species were almost equally present in plaque isolated from healthy denture wearers as well as denture stomatitis patients in this study. Furthermore, the monospecies biofilm formation of these species on denture material discs was evaluated and the resulting model systems were successfully employed to test the antibacterial effectiveness of different denture cleansers.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
We thank Dr. Steven Clegg for providing the K. pneumoniae IA 565 strain and the West China Hospital of Stomatology for providing denture plaque samples. This study was partially supported by a grant (2010-024) from GlaxoSmithKline Consumer Healthcare, Weybridge, UK; the International Science and Technology Cooperation Program of China (2011DFA30940) and the National Basic Research Program of China (2011CB512108), China.
ETHICAL APPROVAL
Ethical Approval was given by the Institutional Review Board at the West China Hospital of Stomatology, China (IRB number: 2012-0004).


