All published articles of this journal are available on ScienceDirect.
HLA-DQB1 Haplotypes and their Relation to Oral Signs Linked to Celiac Disease Diagnosis
Abstract
Objectives:
Celiac disease (CD) is an autoimmune disorder that can be divided into typical and atypical forms. Atypical forms can show extraintestinal manifestations among which oral signs are very frequent. Considering that the pathogenesis of CD is related to a positivity to specific HLA-DQB1 haplotypes, we tested whether the presence of the HLA-DQB1*02 allele could be a hypothetical cause of the development of oral manifestations.
Subjects and Methods:
For this study was been examined the oral condition of 98 Sardinian patients, all affected by CD and all on a gluten-free diet for at least 1 year. Then was been determined each patient’s HLA-DQB1 haplotype and compared these results with clinical information.
Results:
The statistical analysis evidenced that the absence of the HLA-DQB1*02 allele predisposes to oral manifestations such as dental enamel defects (DED) and recurrent aphthous stomatitis (RAS) (Pvalue=5.98x10-05, OR = 0.23, CI: (0.10 - 0.45) per each copy of the HLA allele).
Conclusions:
These results showed that the presence of the HLA-DQB1*02 allele influences the development of oral signs in a dose-dependent manner and also how the HLA haplotype connected to oral signs could have a fundamental role for the diagnosis of atypical forms of CD.
INTRODUCTION
Celiac disease (CD) is an autoimmune disease linked to a permanent sensitivity to gliadin contained in alimentary gluten [1, 2]. This condition determines chronic inflammation on intestinal mucous with consequent parietal injury [3]. The autoimmune response happens as a reaction against tissue transglutaminase autoantigen that principally damages the gastroenteric system determining global malabsorption with progressive flattening of the intestinal villi [4-6].
Celiac disease can be distinguished into different clinical forms known as classical, atypical, subclinical and latent. The classical form is characterised by intestinal symptoms as chronic diarrhea, weight loss, growth deficit, vomiting; all other symptoms from other sites determine the atypical, subclinical and latent forms [7, 8]. Atypical pattern may be often presents only by lesions in the oral mucosa or by defects in dental enamel [9].
CD is strictly linked to a genetic predisposition given by particular human leukocyte antigen (HLA) class II alleles, such as type 1 diabetes, rheumatoid arthritis, multiple sclerosis and Hashimoto’s thyroiditis [10, 11]. Among all HLAcodified antigens, 90% of celiac patients carry the HLA-DQB1*0201 allele (DQ2 antigen) and, if this is absent, the HLA-DQB1*0302 allele (DQ8 antigen) [12, 13]. It is important to highlight that this HLA molecule is almost always cis-encoded by the DRB1*0301-DQA1*0501- DQB1*0201 haplotype, quite common in Sardinia (39% frequency) with one of the world’s highest frequencies [13-16]. In celiac disease we can identify an activation of T cells, linked to HLA expression, in the epithelium and lamina propria of the gut, which disappears after a gluten-free diet. Recent works have shown that T cells do not disappear in the oral mucosa of treated celiac patients due to a still unknown mechanism [17, 18].
HLA typing is an important marker to confirm a diagnosis of CD, moreover today interest is increasing regarding different clinic forms of CD linked to different HLA-DQB1 haplotypes [19].
Among all these clinical manifestations, the prevalence of CD is currently underestimated, since many atypical cases remain undiagnosed [20, 21]. The development of serious or even fatal complications (autoimmunity, cancers) requires the earliest possible diagnosis of CD [22]. This also explains the importance of a strict gluten-free diet for celiac patients who seem to be protected against long-term complications [23, 24].
Oral signs are most frequently CD symptoms, linked to atypical forms, and are represented principally by recurrent aphthous stomatitis (RAS) and dental enamel defects (DED) [6, 9, 25, 26]. The aim of this study is to investigate about a possible correlation between the presence of specific HLA alleles with the presence or absence of RAS and DED.
MATERIALS AND METHODS
Subjects
For this study were analysed 98 celiac patients (23 males and 75 females), with a medium age of 35.92 years (range 7 - 77 years). All patients were typed for anti-gluten antibodies (AGA, both IgA and IgG) and antiendomysial antibody (EMA). Intestinal damage has been graded according to the Marsh classification (type 3, type 2 and type 1). In order to reduce possible confounding effects or intestinal damage due to diseases other than celiac disease, patients with the type 1 and 2 of the classification were excluded before the study. For all participants the diagnosis of CD was stablished by demonstration of enteropathy with type 3 villous atrophy in a small bowel biopsy and all patients had been on a gluten-free diet for at least 1 year. Furthermore, all patients responded to a gluten-free diet, as evaluated upon further regular annual follow-ups.
An anamnestic and diagnostic case study was compiled for each patient to indicate the past or present presence of celiac oral signs, such as RAS and DED [6, 25, 26]. DED were graded 0 to IV according to Aine’s classification [27-29] (Table 1) while RAS linked to CD was determined by the investigation as the past presence of frequently aphtous lesions described by patients as the presence of one or more ulcers at the same time recurring at least 2 times a month, in the period before the gluten-free diet, and never more returned after 1 month of diet. The presence of these lesions was verified by evaluating the medical records of first admission.
Classification of Systemic Enamel Defects in Celiac Disease, (29)
| Grade I | Defects in colour of enamel: single or multiple cream, yellow or brown opacities |
| Grade II | Slight structural defects: rough enamel surface, horizontal grooves, shallow pits |
| Grade III | Evident strucutral defects: deep horizontal grooves, large vertical pits |
| Grade IV | Severe structural defects: shape of the tooth may be changed |
At the time of examination, all participants were informed about the research and its purpose and gave their informed consent in accordance with the ethical standards of the Helsinki Declaration of 1975. Oral brushing was subsequently carried out on all patients, obtaining a sample for DNA extraction allowing the determianation of the HLA-DQB1 haplotype using the conventional PCR method [30].
PCR Technique and Determination of HLA-DQB1 Genotype
To perform the PCR for each sample, a Kit formed from a pre-formed MIX and eight couples of primers was used. Positivity or negativity of amplification performed for each couple allows the HLA-DQB1 genotype to be determined [25-26]. This set of primers will positively identify the HLA-DQB1 alleles corresponding to the serologically defined series HLA-DQ2, DQ3, DQ4, DQ5, DQ6, DQ7, DQ8 and DQ9, thus all combinations of DQB1 can be readily identified. DQ4, DQ5 and DQ6 were uniquely identified, whereas the DQ2 specificity was amplified by three primer mixes, DQ7 and DQ9 specificities were amplified by two primer mixes, DQ3 and DQ8 specificities were amplified by four primer mixes.
On the contrary, on examining the eight primer mixes with corresponding amplified DQB1 alleles, the first primer mix amplified allele group DQB1*05, the second amplified allele group DQB1*06, the third, the fourth and the sixth amplified allele group DQB1*02, the fourth, the fifth, sixth and seventh amplified allele group DQB1*03 and the eighth amplified allele group DQB1*04.
For all the alleles, the reaction was performed in 10.08 μl reaction volumes using the mixture according to the manufacturer's instructions. The mixture contained 3 μl of master mix, 5 μl of DNAsi-RNAsi free water, 0.08 μl of Taq polymerase and 2 μl of DNA suspension, this was put into a tube containing the lyophilized primer pair. The thermocycler profile was as follows: an initial denaturation at 94° C for 2 min; 10 cycles consisting of 94°C for 10 sec and 65° C for 1 min and finally 20 cycles consisting of 94° C for 10 sec, 61° C for 1 min and 72° C for 30 sec. PCR products were analysed by electrophoresis on an agarose gel.
Statistical Analysis
To test the relation between the HLA haplotype and the presence of oral signs, we used a logit regression model where the outcome variable represents the presence (or absence) of the specific oral sign to be tested (DED , RAS, or both). A variable was then coded with values 0, 1 or 2 if the individual carried no, one or two copies of the HLA-DQB1*02 allele, respectively.
RESULTS
Among the 98 patients included in this study, 38.8% (38 persons) were affected by RAS before the gluten-free diet and 28.6% (28 patients) presented DED. Grade I type enamel defects were the only diagnosed. In total 61.2% (60 patients) of the patients showed one or more oral signs (Fig. 1).
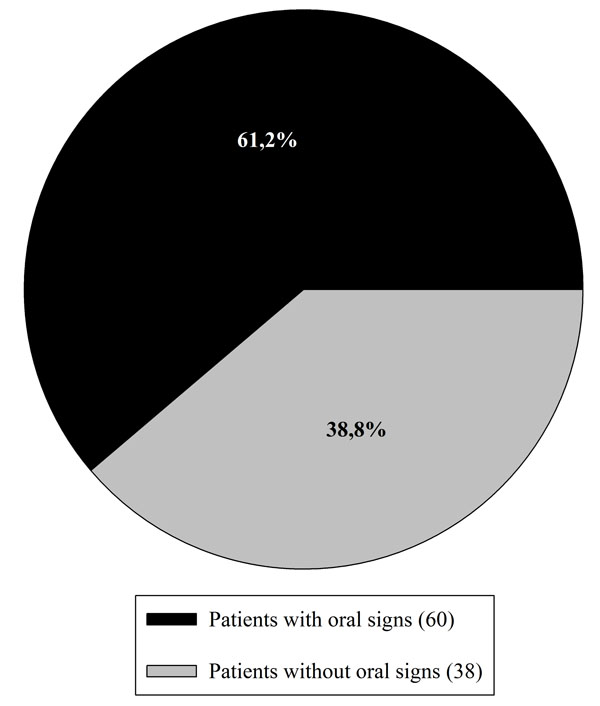
Distribution of DED and RAS among the 98 examined patients.
The majority of the patients carried one (48.0%) or two (33.7%) copies of the HLA-DQB1*02 allele, whereas 18.3% did not carry this allele. Interestingly, we observed that most of the patients carrying one of two copies of the HLA-DQB1*02 allele did not show DED, and this observation was statistically significant (P=0.0003, see Table 2). Similarly, the presence of at least one copy of the HLA allele correlates with a lower frequency of RAS (P=0.002, see Table 2). The statistical evidence was stronger when considering both oral signs. Indeed, all individuals who did not carry a copy of the HLA-DQB1*02 allele presented RAS or DED (P=5.98x10-5, see Table 2 and Fig. 2), while no appreciable difference was observed between those carrying one or two copies.
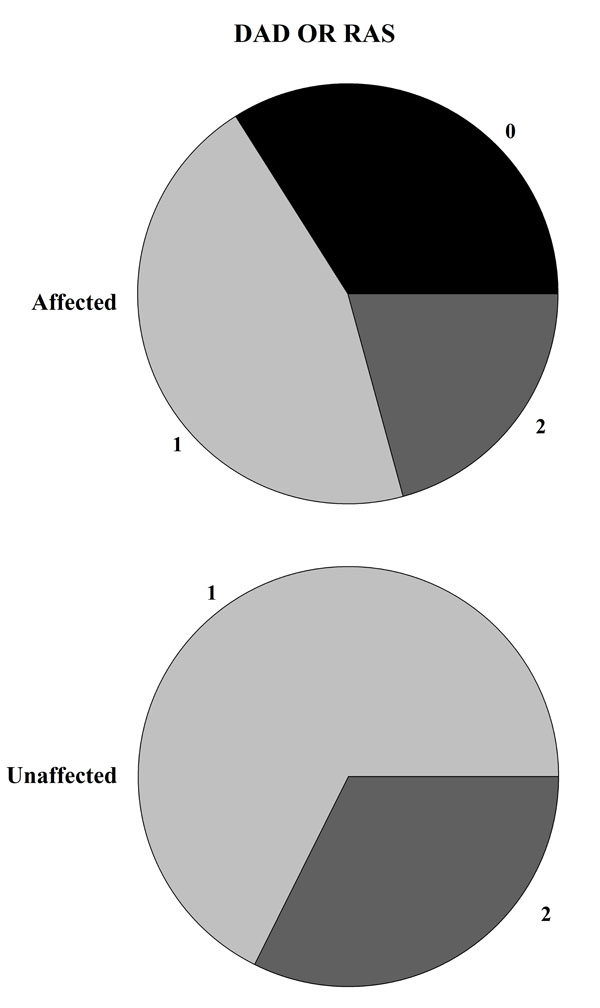
Visual representation of HLA-DBQ*02 haplotype distribution (2 = Homozygosis, 1 = Eterozygosis, 0 = allele’s absence) between individuals affected or unaffected by DED or RAS. If the DQ2 antigen is absent the 100% of patients show at least one signs between DED and RAS.
This Table Summarizes the Distribution of the HLA-DBQ*02 Haplotype Among Individuals Affected by DED, or RAS, or by One of these Two. For Each Oral Sign Tested, We Reported the p-Value and the Odds Ratio for Each Copy of the HLA Allele
| Copies of the HLA-DBQ*02 | N Affected | N Unaffected | Affected% | Unaffected% | OR (CI -2.5-97.5) | Pvalue |
|---|---|---|---|---|---|---|
| DED | ||||||
| 0 | 12 | 6 | 0.428 | 0.085 | ||
| 1 | 12 | 35 | 0.428 | 0.500 | ||
| 2 | 4 | 29 | 0.142 | 0.414 | ||
| 0.25 ( 0.11-0.51) | 0.0003 | |||||
| RAS | ||||||
| 0 | 12 | 6 | 0.315 | 0.100 | ||
| 1 | 19 | 28 | 0.500 | 0.467 | ||
| 2 | 7 | 26 | 0.185 | 0.433 | ||
| 0.37 (0.18 -0.68) | 0.002 | |||||
| DED or RAS | ||||||
| 0 | 18 | 0 | 0.340 | 0.000 | ||
| 1 | 24 | 23 | 0.453 | 0.676 | ||
| 2 | 11 | 22 | 0.207 | 0.324 | ||
| 0.23 (0.10- 0.45) | 5.98x10-5 | |||||
DISCUSSION
For modern medicine, genetic screening is becoming more important thanks to major advances in research into pathology etiology. Nowadays HLA is one of the most studied genetic loci because it is linked to many genetic diseases; among these,celiac disease is an important candidate for public health newborn genetic screening based on HLA-DQ alleles [31].
Genetic screening on HLA locus linked to celiac development will primarily eliminate the further need for the 60% of the population lacking the DQ2 and DQ8 antigens to undergo serial autoantibody testing but the aim of this work was to show how HLA could be important not just for obtaining an “exclusion diagnosis” [32, 33].
In recent years, the increasing detection of atypical forms of CD, has made it necessary to find more precise early diagnostic methods that permit the underlying part of the “celiac iceberg "to be discovered [21, 34]. In this study on 98 CD patients from Sardinia we evaluated the possible relationship between the HLA haplotype and the development of oral diseases to verify the value of these oral signs as diagnostic instruments of CD. We observed that the absence of the HLA-DQB1*02 allele is associated with the presence of at least RAS or DED, while no appreciable difference was observed between those carrying one or two copies. To explain these results, anaylsis on the binding capacity to the pocket of HLA-DQ2 or HLA-DQ8 molecules on antigen-presenting cells [35] must be carried out. CD immunitary reactions are linked to the relationship established by this complex and CD4+ T cells causing tissue damage in the intestinal mucosa. This tissue damage seems to be correlated to an increasing presentation of gluten epitopes to T cells associated to the presence of the HLA heterodimer DQB1*02 which differ in their peptide binding affinity for gluten peptides [36]. This data explain why DQ2 homozygosis in intestinal mucosa is linked to a more marked intestinal flattening of the villi [13]; faced with these results we need to understand why the oral mucosa seem to have opposite behaviour.
This different response could be explained by the immunological and histological differences between the small intestine and the gut. The pathological alterations linked to celiac disease are correlated to T-cell activity in the presence of gliadin-derived peptide. This mechanism is well studied in the small intestine, where lymphocytes induce mucosal damage while the exact pathogenetic mechanism that leads to the development of lesions in the oral mucosa is still unknown.
There are many hypotheses about the different responses of intestinal or oral mucosa. The existence of different gliadin peptides that could provoke an immunological reaction depending on different anatomic or histological areas, the activity of oral bacteria as first participants in the enzymatic degradation of gliadin or the histological differences between the intestinal mucosal epithelium, consisting of single layered cuboidal cells compared to the stratified epithelium of the oral mucosa, which could explain the different concentration of gliadin-derived peptide [37, 38]. More accurate estimation and confirmation of the effect of specific HLA alleles require further evaluation in a larger sampling paying particular attention to the possible interaction of HLA loci with other genes (gene-gene interaction) [39]. Any possible further study could also focus on the application of more rapid, though more expensive, methods to obtain the HLA haplotype such as PCR-realtime [40].
Overall, our results suggest that genetic predisposition at the HLA-DQB1 locus, influences the formation of celiac oral signs in CD and could become a fundamental test for recognising underlying celiac forms. In conclusion, further studies will be necessary to establish if the role of oral pathology can be significant in identifying latent and atypical forms of celiac disease.
CONFLICT OF INTEREST
None Declare.
ACKNOWLEDGEMENTS
The authors received no specific funding for this article, and declare that no competing interests exist.


