All published articles of this journal are available on ScienceDirect.
Anti-Quorum Sensing Effect of Salvadora Persica Against Enterococcus faecalis (ATCC 29212)
Abstract
Introduction:
Quorum Sensing (QS) is a mechanism many bacteria use to manage their cooperative activities and physiological functions. The Fsr system in Enterococcus faecalis (ATCC 29,212) is an example of quorum sensing with a cell density-dependent two-component regulatory system mechanism. Several publications have shown that the Fsr system and proteases independently contribute to E. faecalis pathogenicity in various infection models.
Objectives:
There is currently no published research to determine the exact molecular ability of Salvadora persica on quorum-sensing genes. Therefore, this study aimed to determine the plant extracts that inhibit the expression of the quorum-sensing gene (FsrC).
Methods:
Different fractions of Salvadora persica were obtained using different solvents, including standard hexane, chloroform, ethyl acetate, n-butanol, ethyl alcohol, and water which are expressed as fractions 1,2,3,4,5 and 6, respectively. Antibacterial activity assay of different plant extracts (S. persica) was determined by minimum inhibitory concentration (MIC). Finally, the relative expression of the quorum-sensing (QS) gene was evaluated using a One-step quantitative RT-PCR PrimeScript™ RT-PCR Kit.
Results:
All fractions of S. persica showed antimicrobial activity. However, ethyl acetate- S. persica inhibited the growth of E. faecalis (ATCC 29,212) at the lowest concentration, which was 20mg/ml and the highest concentration inhibited the growth of E. faecalis (ATCC 29,212) was 60mg/ml (chloroform- S. persica). Furthermore, the highest change fold value of (4.99) was recorded in treated E. faecalis (ATCC 29,212) with fraction 1 (hexane).
Conclusion:
Overall, S. persica showed antimicrobial activity against E. faecalis (ATCC 29,212). However, more studies are required to investigate the effect of different plant extracts on quorum-sensing genes of Enterococcus faecalis.
1. INTRODUCTION
The most prevalent enterococcal species found in clinical and dietary samples are Enterococcus faecalis and Enterococcus faecium, which are responsible for around 90% and 10% of human nosocomial infections, respectively [1-6].
Quorum sensing (QS) is a mechanism used by many bacteria to manage their cooperative activities and physiological functions [7, 8]. Quorum sensing (QS) is a population-based control based on self-secreted molecules known as auto-inducers (AI); they can accumulate in the environment and change the expression of many genes [9]. Pathogenicity, biofilm formation, motility, sporulation, conjugal plasmid transfer, bioluminescence, antibiotic resistance, and antibiotic synthesis are all regulated by quorum sensing (QS) [10, 11]. Generally, the QS systems in bacteria are categorised into at least three classes, including LuxI/LuxR-type QS in Gram-negative bacteria, oligopeptides two-component-type QS in Gram-positive bacteria, and (3) luxS-encoded autoinducer 2 (AI-2) QS in both Gram-negative and Gram-positive bacteria [11, 12]. The Fsr system in E. faecalis is an example of QS with a cell density-dependent two-component regulatory system mechanism [13]. The fsr locus is 2.8 kb in size and comprises four genes: fsrA, fsrB, fsrD, and fsrC [8, 14] and the Fsr system is required for four genes including gelE, sprE, ef1097, and ef1097b [15]. Fsrc encodes the transmembrane histidine protein kinase FsrC, the sensor-transmitter of the fsr operon [14]. However, the Fsr system has been identified as a quorum-sensing locus that responds to the extracellular accumulation of a peptide lactone encoded at the C terminus of the FsrB protein [14]. The FsrC histidine kinase is presumably triggered by the accumulation of the peptide in the extracellular space, resulting in the activation of the response regulator and transcription factor FsrA [16].
Several publications have shown that both Fsr and proteases independently contribute to E. faecalis pathogenicity in various infection models [17-22]. The specific mechanisms through which Fsr and its regulatory proteases contribute to infection toxicity are unknown [15]. As mentioned previously, the accumulation of these molecules changes the expression of many genes, including the genes responsible for forming biofilm [23]. A previous study revealed that quorum-sensing is significant in biofilms and may result in dispersion [24]. Antibiotics are used to treat various infectious illnesses [25]. However, the overuse of antibiotics resulted in drug-resistant bacterial strains, including (E. faecalis) [26]. There is a great demand for new agents and natural compounds because of low effectiveness due to the development of resistance, host toxicity, and various side effects of currently used therapeutics [27]. As a result, blocking bacterial quorum-sensing (QS) systems have gotten a lot of attention since it has the potential to reduce bacterial virulence without causing bactericidal pressure, which can lead to drug resistance. Actinobacteria continue to produce novel molecules with various bioactive properties, such as antibacterial, anticancer, and antihelminthic properties as well as the emergence of integrative metabolomic and genomic analysis has provided new hope for natural product researchers [28]. Recent study confirmed that Pantoea Natural Product 3 (PNP-3) is a broad-spectrum antimicrobial that is effective against a variety of multidrug-resistant bacteria [29]. Discovering novel natural product with antimicrobial activity is critical as drug-resistant bacteria have few therapeutic options.
S. persica (Miswak-Siwak) is a Middle Eastern native bushy plant whose parts have been used as a natural toothbrush and mouth rinse from time immemorial [30, 31]. However, there is currently no published research to determine the exact molecular capability of S. persica to QS genes. Therefore, this study aimed to determine the plant extracts (S. persica) that inhibit the expression of the QS gene (FsrC).
2. MATERIALS AND METHODS
2.1. Bacterial Culture
Enterococcus faecalis (ATCC 29,212) were used in all experiments obtained from the American Type Culture Collection (ATCC), cultured out twice on BHI (Scharlau Chemie S.A., Barcelona, Spain) agar plates for 24 hours at 37ºC and checked for purity by Gram stain and colony morphology [32].
2.2. Plant Extracts
Different solvents were used to extract active substances from plants; the solvents are selected primarily based on the polarity of the solute, from low polar to high polar, as follows: Hexane – Chloroform – Ethylacetate – Butanol – Ethanol – Water [33]. All solvents used for the extraction process were used as blanks and controls in all experiments throughout the study to exclude the effect of the solvents. The resultant liquid was filtered through a filter paper into a round-bottom flask and concentrated using a Buchi R-200 rotary evaporator, thus removing all the solvents and resulting in a viscous crude plant extract. The crude extracts were stored at - 20°C and freshly dissolved in 10% DMSO before use.
The mixture was prepared from 50g of dried Miskwak (S. persica), and 100ml of Hexane put in an ultrasonic bath (1.5 hours at 37°C). Then the mixture was filtrated to get extract part one (fraction 1) solvent removed using the rotary evaporator. The residue then dried at room temperature and was used for the next solvent extraction. The same technique was used for the other solvents to get the different extract fractions (fraction 2 from chloroform, fraction 3 from ethyl acetate, fraction 4 from butanol, fraction 5 from ethanol, and fraction 6 from water).
2.3. Determination of Minimum Inhibitory Concentration (MIC)
The minimum inhibitory concentration (MIC) for antibacterial activity assay of different plant extracts was determined by serial dilution, as described by Eloff [34], according to the Clinical and Laboratory Standards Institute (CLSI) standard protocol [35]. In 1.5 ml microcentrifuge tubes (Eppendorf), stock solutions of the respective plant extracts were prepared by dissolving the dry plant extract in dimethylsulphoxide (DMSO) to a final concentration of 100mg/ml. The serial dilutions from the stock solution were made in 2ml-96-well microplates to the concentration of (5, 10, 20, 25, 30, 35, 40, 50, 60, 70 and 100 mg/ml). Müeller-Hinton broth was used to obtain an activated bacterial suspension with a concentration of 1.5 x 108 CFU/ml (0.5 MacFarland). 100µl of bacterial suspension was transferred into each well and inoculated at 37°C for 24 hours to activate. The growth control and sterile control were used for this assay; growth control contained only bacterial suspension, and sterile control contained only plant extracts. The effect of solvents on the bacterial growth was not recorded as a stock solution prepared from the dry crude extract and 10% DMSO solution. MIC was determined by reading the absorbency at wavelength 590 nm, and the lowest concentration of each extract displaying no visible growth was recorded as the minimum inhibitory concentration. All samples were tested in triplicate.
2.4. RNA Extraction and Preparation
Total RNA was isolated by RNXplus kit (SinaClon, Tehran, Iran) from untreated and treated samples E. faecalis (ATCC 29,212). The RNA from treated samples was extracted after determining the minimum inhibitory concentration (MIC). The bacterial culture with the highest plant concentration that showed visible growth (a concentration before MIC) was selected for the RNA extract and considered a treated sample. Briefly, bacterial cells were dissolved in 1ml ice-cold RNXplus solution, gently shacked after adding 200 ml chloroform, and then kept on ice for 15 minutes, followed by cold centrifugation at 12000rpm for 15 minutes. Wqualvolume of isopropanol was added, followed by cold centrifugation at 12000 rpm for 15 minutes. The supernatant was discarded, 1ml Ethanol (75%) was added, and then centrifuged at 7500 rpm for 8 minutes. The supernatant was removed, and the pellet was left to dry at room temperature. Finally, it dissolved in 50μl DEPC (Diethylpyrocarbonate) (SinaClon) water. RNase-free DNase was used to eliminate DNA from the RNA fraction. Additionally, Nanodrop (BioTech, USA) was used to determine the quality of isolated RNA and immediately quantified by RT-PCR.
2.5. Quantitative RT-PCR Assay
The nucleic acid sequence of the QS gene (FsrC) was obtained from Enterococcus faecalis (ATCC 29,212). Forward primer (GCTTATTTGGAAGAACAACGTATCAA) and reverse primer (CGAAACATCGCTAGCTCTTCGT) were selected to evaluate the expression of the target QS gene [36]. Following the manufacturer's instructions, the relative expression of the QS gene (FsrC) was evaluated using a One-step quantitative RT-PCR PrimeScript™ RT-PCR Kit. The total volume was 20 μl, which contained 1 μl of each primer sequence, 2 μl of 2X reaction buffer, 1 μl of the enzyme solution, and 3 μl of extracted RNA. The reaction was carried out at 50°C for 20 minutes (cDNA synthesis), with the initial activation step at 94°C for 5 minutes, followed by 40 cycles of denaturation at 94°C for 25 seconds, annealing at 63°C for 60 seconds, and extension at 68°C for 60 seconds. All results were normalized by untreated samples. The effect of plant extracts on the quorum-sensing gene was determined using comparative critical threshold (Ct) real-time PCR. The following formula (ΔCq= Cq treated bacteria – Cq control bacteria) and normalize= 2-ΔCq are used to determine the change fold and normalize change value. All experiments were performed twice.
3. RESULTS
3.1. Minimum Inhibitory Concentration (MIC)
All fractions of S. persica showed antimicrobial activity, as shown in Table 1 and Fig. (1). However, ethyl acetate- S. persica inhibited the growth of E. faecalis at the lowest concentration, which was 20mg/ml, and the highest concentration, which was 60mg/ml (chloroform- S. persica).
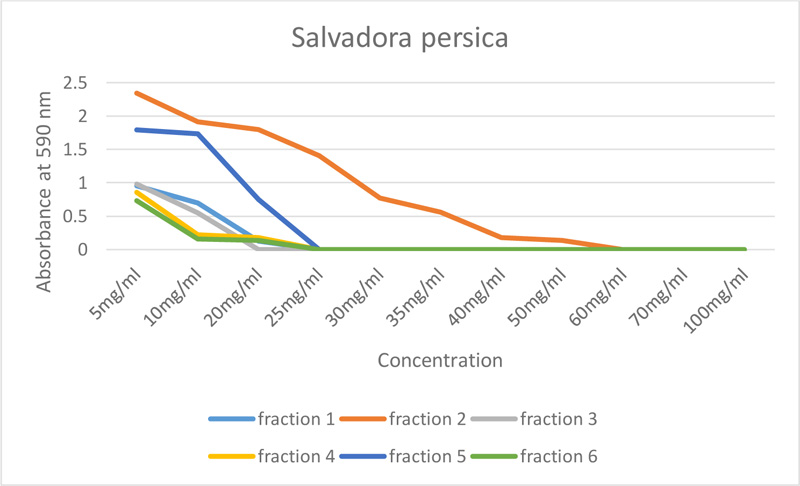
| S. persica | 5mg/ml | 10mg/ml | 20mg/ml | 25mg/ml | 30mg/ml | 35mg/ml | 40mg/ml | 50mg/ml | 60mg/ml | 70mg/ml | 100mg/ml |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fraction 1 | 0.955 | 0.702 | 0.131 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fraction 2 | 2.343 | 1.911 | 1.802 | 1.408 | 0.777 | 0.563 | 0.183 | 0.142 | 0 | 0 | 0 |
| Fraction 3 | 0.981 | 0.546 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fraction 4 | 0.857 | 0.220 | 0.181 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fraction 5 | 1.793 | 1.734 | 0.753 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fraction 6 | 0.732 | 0.158 | 0.134 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Parameter | fraction 1 | fraction 2 | fraction 3 | fraction 4 | fraction 5 | fraction 6 |
|---|---|---|---|---|---|---|
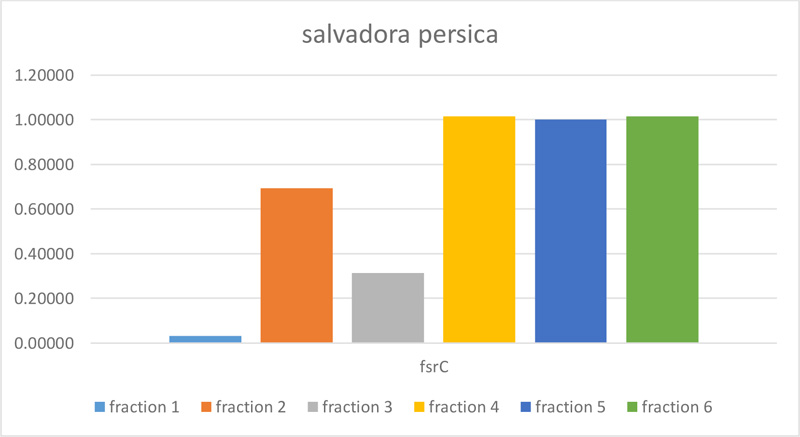
| normalize change value | 0.03147 | 0.69255 | 0.31208 | 1.01396 | 1.00000 | 1.01396 |
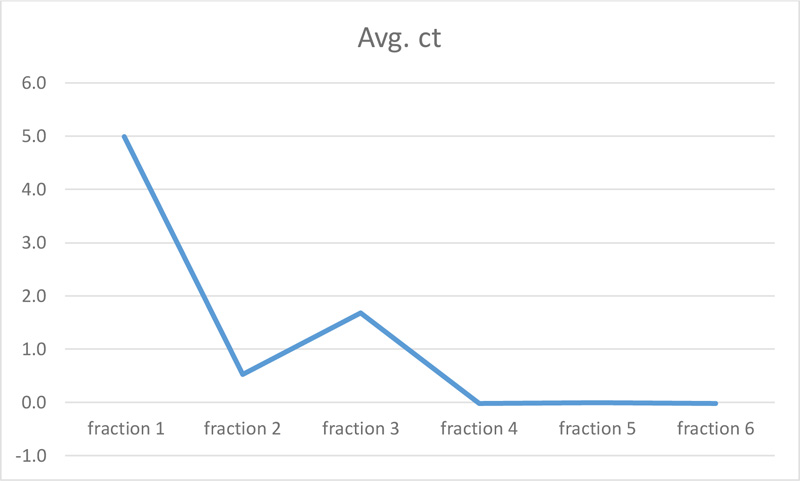
| Change fold | 4.99 | 0.53 | 1.68 | -0.02 | 0.00 | -0.02 |
3.2. Effect of S. Persica on the Expression of QS FsrC Gene
The molecular effect of S. persica extract against E. faecalis (ATCC 29,212) was determined by evaluating the relative expression levels of FsrC genes. For this purpose, Real-time quantitative polymerase chain reaction (RT-qPCR) was used. As shown in Table 2 and Figs. (2 and 3), fractions 4, 5, and 6 of S. persica showed no effect on the level of FsrC expression, while the expression of the same gene decreased in bacterial samples that were treated with fractions 1, 2 and 3 of S. persica. Furthermore, the highest change fold value of (4.99) was recorded in treated E. faecalis (ATCC 29,212) with fraction 1, as shown in Fig. (3).
4. DISCUSSION
Currently, E. faecalis is widely known as one of the most common bacteria in the root canal [12, 37]. On the other hand, several investigations reported that both Fsr and proteases independently contribute to E. faecalis pathogenicity in various infection models [17-20, 22]. The QS system encoded by the fsr gene cluster on the E. faecalis chromosome regulates the production of two pathogenicity-related proteases, gelatinase (GelE) and serine protease (SprE) [25]. Moreover, deletion of fsrA, fsrB, or fsrC inhibits the expression of gelE and sprE in wild-type E. faecalis strain OG1RF; nonetheless, both were strongly expressed in wild-type E. faecalis strain OG1RF [38].
The rising incidence of antibiotic resistance by pathogenic microorganisms is an alarming issue in treating diseases. There is a significant rise in vancomycin-resistant E. faecalis, which poses a threat considering that this bacterial species already inherently harbor natural antibiotic-resistant characteristics [36, 39]. Thus, continuous search and discovery of new therapies would hopefully replace those antibiotics to which bacteria may develop resistance. Recently, a new approach has been identified by detecting Anti-QS compounds of herbal origin [40, 41]. Halogenated furanones produced by the benthic marine macroalga Delisea pulchra were the first identified anti-QS compounds [42]. Anti-QS compounds are known to have the ability to prohibit bacterial pathogenicity [43]. There is growing interest in implementing novel strategies based on natural compounds to eradicate bacteria, including phytochemicals with anti-biofilm, anti-QS, and anti-oxidant properties that may enhance the efficacy of antibiotics, allowing to reduce their use [44].
S. persica (Miswak-Siwak) is a Middle Eastern native bushy plant whose parts have been used as a natural toothbrush and mouth rinse from time immemorial [30]. Extracts of S. persica have even been put into mouthwashes for oral hygiene as it contains trimethylamine, salvadorime chloride, and fluoride in high amounts [45, 46]. S. persica is being studied for its antibacterial action against Streptococcus mutant and E. faecalis [46-48]. Other studies reported that S. persica has an antibacterial action, allowing it to be used as an irrigant solution in endodontic treatment against endodontic infections, and suggested that it can be used as a root canal irrigant substitute for sodium hypochlorite and chlorhexidine [49, 50]. However, there is currently no published research to determine the exact molecular capability of S. persica on QS genes. The present study clearly determined the effects of S. persica extracts with different solvents on E. faecalis through evaluating the expression of target QS gene (FsrC).
In this study, the antibacterial activity of S. Persica extract solution was assessed in vitro using the broth microdilution method as plant samples are difficult to diffuse in the medium [51, 52]. Previous investigations in Iraq reported that microorganisms in the root canal are highly sensitive to the alcoholic extract of S. Persica solution at 15% [49]. The present study also confirms that E. faecalis (ATCC 29,212) is highly sensitive to all mentioned fractions of S. persica. The antimicrobial effect of S. Persica is believed to be due to its high chemical contents of chlorides, tannins, trimethylamine, salvadorine, nitrate, thiocyanate and sulphur [53]. This study confirmed that S. persica decreased the expression level of QS gene in E. faecalis (ATCC 29,212) and it had antibacterial properties. This finding agrees with previous studies that suggested the Anti-QS ability of S. persica against E. faecalis [30]. In general, all AQS compounds have a wide range of actions that target practically all bacterial species that depend on QS pathways [10, 11]. This suggests that AQS in S. persica may also function against other opportunistic oral pathogens. Therefore, more studies are required to investigate the AQS ability of S. persica against other oral pathogens.
CONCLUSION
The present study concluded that fractions 1, 2, and 3 decreased the expression of the QS gene (FsrC), and the highest change fold value (4.99) was recorded by hexane-S. persica extracts. Moreover, the lowest concentration of S. persica that inhibited the growth of E. faecalis was 20mg/ml of fraction 3 (ethyl acetate- S. persica). Overall the findings suggest that S. persica showed antimicrobial activity against E. faecalis.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No humans or animals were used in this study.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The corresponding author confirms that ethical considerations were taken for this research, the article is original, and its contents are unpublished. The co-author has read and approved the manuscript for submission.


