All published articles of this journal are available on ScienceDirect.
Comparison of the Expression of Periodontal Markers in Dental and Bone Marrow-derived Mesenchymal Stem Cells.
Abstract
Background:
Periodontal ligament stem cells are a source of mesenchymal stem cells, but it is unclear whether their phenotype is distinct from mesenchymal stem cells derived from different tissues, such as those derived from bone marrow.
Objective:
To investigate the expression of the putative PDL markers asporin, periostin, nestin and cementum protein 1, by periodontal ligament stem cells both constitutively and during osteogenic differentiation when compared to bone marrow-derived mesenchymal stem cells, and dental pulp stem cells.
Methods:
The primary human periodontal ligament, bone marrow, and dental pulp stem cells, and osteoblasts from different donors were cultured in vitro. The expression of periodontal marker associated genes during osteogenic induction was tested by qRT-PCR and immunofluorescence staining.
Results:
Asporin expression was detected in periodontal ligament stem cells and increased markedly during the time in culture (upregulated x53 fold at 21 days post-induction). During osteogenic differentiation, asporin expression significantly decreased in periodontal ligament cells whereas periostin significantly decreased in dental pulp cells. Periostin expression was absent in osteoblasts, but expression gradually increased in all other cells with time in culture. Nestin expression was mainly seen in the periodontal ligament and dental pulp cells and was largely absent in osteoblasts and bone marrow cells. Cementum protein-1 was most highly expressed in bone marrow cells and osteoblasts following osteogenic induction.
Conclusions:
The results provide further evidence that periodontal ligament-derived and bone marrow derived mesenchymal stem cells are phenotypically distinct. Periodontal markers are also expressed in dental pulp stem cells.
1. INTRODUCTION
Mesenchymal Stem Cells (MSCs) are adult stem cells that exhibit self-renewal and have the capacity to differentiate into osteoblast, chondrocyte, and adipocytic lineages. They were originally isolated from the stroma of bone marrow [1] but since then have also been isolated from many other tissues. Within the dental tissues MSCs have been isolated from Periodontal Ligament (PDL), dental pulp, dental follicle, exfoliated teeth (SHED cells) and gingival connective tissues [2].
There is considerable interest in the potential therapeutic application of MSCs for tissue regeneration, including for periodontal regeneration. A number of animal studies have demonstrated the principle of periodontal regeneration using MSCs derived from sources including bone marrow, PDL, gingiva, dental pulp and adipose tissue [3-8]. However, the question remains whether these different MSC sources are phenotypically distinct, which might render them particularly suitable for a specific regenerative application such as periodontal regeneration. A few studies have suggested that PDL-derived MSC may have phenotypic differences with other MSCs [9-11]. Studies to address this question are hampered by the absence of any specific markers for either PDL cells or tissues. Nevertheless, a number of proteins are reported to be highly expressed in PDL and have been proposed as PDL markers in studies investigating differences between MSC phenotypes, even though they may not be uniquely expressed in that tissue only. These putative candidate PDL markers include periostin, asporin (periodontal ligament associated protein-1, PLAP-1), cementum protein-1 (CEMP-1) and the neural crest-derived tissue marker Nestin [12].
Asporin is a member of the small leucine-rich proteoglycan family and was originally described as being localised to the periodontal ligament, although it is expressed in a number of other tissues [13].
Periostin is a matricellular protein involved with the maintenance of connective tissue integrity which is known to be highly expressed in the periodontal ligament, as well as in the connective tissues including bone.
Nestin is an Intermediate Filament (IF) protein, which interacts with other IF proteins in the cytoskeleton. Nestin expression is characteristic of neural crest-derived cells, particularly when undifferentiated [14, 15].
Cementum-protein-1 (CEMP-1) was first isolated from a cementoblastoma as a potentially cementum-specific protein. CEMP-1 expression is seen in cementoblasts but has also been reported in bone [16]. CEMP-1 reportedly has growth factor-like activity on PDL cells and other undifferentiated mesenchymal cells stimulating osteoblast/cementoblast differentiation.
Consequently, the aim of the study here is to test the hypothesis that PDL-derived MSC are phenotypically distinct from MSCs derived from other sources. Specifically, the aim was to test the hypothesis that PDL-derived MSC (PDLSC) will differentially express high levels of periostin, asporin, CEMP-1, and nestin when compared to expression in dental pulp-derived MSC (DPSC) and bone marrow-derived MSCs (BMSCs).
2. MATERIALS AND METHODS
2.1. Overview
PDLSC and BMSC derived from 3 different donors along with DPSC from 2 different donors were tested for the expression of marker genes. Primary osteoblast cultures were also tested as control cultures. Cells were first tested for their MSC properties by the capacity to undergo multilineage differentiation and expression of MSC-associated cell surface markers. Gene expression of aspirin (ASPN), periostin (POSTN), nestin (NES) and cementum protein-1 (CEMP1) were tested in cell lines at time points up to 21 days, in normal culture medium and osteogenic medium. Protein expression was confirmed by immunofluorescence.
2.2. Cell Culture
Periodontal ligament cell cultures were established from explants derived from healthy third molar teeth extracted from three patients aged between 20-35 years old (Arceo 1991). Ethical approval for obtaining the samples was obtained from Kent NHS Research Ethics Committee (Reference No: 11/LO/0259). Primary human dental pulp stem cell cultures (DPSC) and human osteoblast cultures derived from alveolar bone and femur were kindly provided by Professor Lucy Di-Silvio, King’s College London. Primary human bone marrow stromal MSCs (BMSC) were purchased commercially (Lonza, Slough, UK). The detailed phenotypic features of the three specific BMSC lines have been described extensively in a previous publication from our group [17]. Cells were maintained in α-Minimal Essential Medium (MEM), penicillin 50 U/ml, streptomycin 50 μg/ml (Sigma–Aldrich), Glutamax 2 mM (Invitrogen, Paisley, UK), and 10% fetal bovine serum/FBS (Sigma–Aldrich) (normal growth medium) at 37°C in a humidified 5% CO2, 95% air atmosphere. For experiments involving the stimulation of osteogenic differentiation, normal growth medium was supplemented additionally with 0.1 μM dexamethasone, 0.05 mM ascorbic acid, and 10 mM ß-glycerophosphate (Sigma–Aldrich). For adipocytic differentiation, normal growth medium was supplemented with 1 μM dexamethasone, 0.25 mM isobutylmethylxanthine, 50 μM indomethacin and 10 μg/ml insulin (Sigma–Aldrich) and for chondrocytic differentiation with 1 μM dexamethasone, 1 μM ascorbic acid, 1% sodium pyruvate (Invitrogen) and 10ng/ml transforming growth factor-beta 1 (TGF-β1, Peptrotech, Rocky Hill, NJ, USA).
All cell types were used in the same passage in each experiment (passage 3-7). Differentiation assays were carried out as previously described [17].
2.3. Flow Cytometry
MSC surface marker expression was measured using four-colour flow cytometry analysis, with the FACSCanto II flow cytometer (BD Biosciences, San Jose, CA). Briefly, 100,000 cells were incubated with Fc block (BioLegend, Cambridge, U.K.) in 50μl of fluorescence-activated cell sorting (FACS) buffer for 15 minutes at room temperature. The antibody cocktail was divided into two groups (A&B). Group A contained CD34-PerCP/Cy5.5 (clone 4H11), CD105-fluorescein isothiocyanate (FITC) (clone 43A3), CD146-phycoerythrin, and (PE)/Cy7 (clone SHM-57) whereas group B contained CD45-PerCP/Cy5.5 (clone HI30), CD44-FITC (clone BJ18), and CD90-allophycocyanin (APC) (clone 5E10). Adding an antibody cocktail into the FACS tube, which contained the cells suspension, promoted direct binding.
Unstained cells, compensations beads, and fluorescence minus one (FMO) were used to set the gating. All antibodies were obtained from Bio-Legend U.K. (Cambridge Bioscience) and used at the recommended concentration. Cells were incubated for 15 minutes at room temperature in the dark. After being washed twice, cells were resuspended in 300μl of FACS buffer and immediately being analysed. A total of 10,000 events were acquired for each sample and data analysis was performed using DIVA software (BD Biosciences).
2.4. Real Time PCR
Total RNA was extracted using TRI reagent (Ambion, Warrington, UK). Cells in 12-well plates were lysed in 600ml TRI reagent and transferred into Phase Lock Gel Heavy tubes (5 prime, VWR, Leicestershire, UK) according to the manufacturer’s instructions. RNA purity and quantity were assessed by Nano Drop 1000 Mass spectrometer (Fisher Scientific). Ratios A260/A280 between 1.8–2 were considered to be high purity and used for cDNA synthesis. Possible DNA contamination was removed and cDNA prepared from 1 μg RNA using QuantiTect Reverse Transcription Kit (Qiagen, West Sussex, UK) according to manufacturer’s instructions. qRT-PCR was performed with a Rotor-Gene 6000 thermal cycler (Qiagen) using Brilliant III Ultra-Fast SYBR Green qPCR Master mix (Stratagene, Agilent Technologies, Cheshire, UK). Primer pair sequences are listed in Table 1. PCR conditions consisted of 1 cycle of 95°C for 3 min, 40 cycles of 95°C for 10 sec, 60°C for 10 sec, and followed by melting analysis of 1 cycle with gradual increase from 65°C to 95°C. RPL13a was used as the reference gene
| Gene | Gene Acc | Forward 5’-3’ | Reverse 5’-3’ |
|---|---|---|---|
| RPL13a | NM 000977 | CATCGTGGCTAAACAGGTACTG | GCACGACCTTGAGGGCAGCC |
| ASPN | SY 130414037 | CCCTTCAGGATTACCAGAGTTG | TTGGCACTGTTGGACAGAAG |
| CEMP1 | SY 140609461 | CCTGGGGATGTGTGAAGAGA | TGTTTTCTGCTCCCTGGACT |
| NES | SY 140307726 | CTCCAAGACTTCCCTCAGCT | TCAGGACTGGGAGCAAAGAT |
| POSTN | SY 130414039 | TGACACAACCTGGAGACTGG | GAGCATTTTTGTCCCGTATCA |
2.5. Analysis of qRT PCR Data
To calculate the relative expression of genes, all samples were normalized to their housekeeping gene (RPL) cycle threshold (Ct) value and then normalised to the average of PDLSC Ct value at day 0 (24 hours after seeding the cells). Thus normalised data from each different cell line (of the same derivation) were combined for analyses. The fold change in gene expression was assessed using the comparative method. Data were expressed as mean ± S.D. Either one-way, or two-way, ANOVA with Bonferroni post test was used to determine whether differences in gene expression levels in the different cell types were statistically significant from control. Statistical analysis was carried out using Prism 5 v5.0 software (Graph Pad Software Inc.). The statistical significance was set at p<0.05.
2.6. Immunofluorescence Staining
Cells were seeded in normal media and at day 3 cells were fixed for 5 minutes with 100% methanol for asporin or periostin staining, and 10 minutes 4% formaldehyde fixed for nestin staining. Thereafter the cells were incubated in 1%BSA / 10% normal goat serum / 0.3M glycine in 0.1% PBS-Tween for 1 hour to permeabilise the cells and block non-specific protein-protein interactions. The cells were then incubated with the antibody asporin (ab58741, 5µg/ml), periostin (ab14041, 1/1000 dilution) and nestin (ab 105389,1/200 dilution) overnight at +4°C. The primary antibody was visualized using Alexa Fluor® 555-conjugated secondary antibodies anti-rabbit IgG (H+L),F(ab’2) (Cell Signaling #4413) at a 1/250 dilution for 1 hour . DAPI was used to counterstain the cell nuclei (blue) at a concentration of 1.43µM. As negative controls, some samples were incubated without the primary antibody. Analysis particle from ImageJ was used to determine the integrated density as a semi-quantitative estimate of protein expression corrected for cell number. This program measures the cell numbers from the DAPI nuclear counter stain, and the total positive fluorescence area in each image.
3. RESULTS
3.1. Stem Cell Characteristics of Cells
Flow cytometry demonstrated that a very high proportion of PDLSC and BMSC cells expressed CD44, CD90, CD146, CD105, and CD45 and did not express the hematopoietic cell markers CD34 and CD45. DPSCs also showed a similar expression pattern of surface markers except that CD146 was only expressed in 72% of cells, and CD105 expression was particularly low with only 11% of cells (Table 2). (NB the data for BMSC shown here was obtained in a previously published study from our group [17].
| - | N | CD44 | CD146 | CD90 | CD105 | CD45 | CD34 |
|---|---|---|---|---|---|---|---|
| PDLSC | 3 | 99.3% ± 1.1 | 95.4% ± 3.9 | 99.4% ± 0.7 | 86.4% + 10.9 | 0.8% ± 0.8 | 0.7% +0.9 |
| DPSC | 2 | 99.8% ± 0.1 | 72.3% ± 15.1 | 97.1% + 0.2 | 11.7% + 2.1 | 0.5% ± 0.7 | 0.5% ± 0.2 |
| BMSC | 3 | 95.1% ± 4.4 | 91.8% ± 4.6 | 96.7% ± 6.4 | 95.6% ± 3.8 | 1.2% ± 1.2 | 2.0% ± 0.7 |
PDLSCs, DPSCs, and BMSCs were able to differentiate into osteoblast, adipocyte, and chondrocyte lineages following 21 days in osteogenic, adipogenic or chondrogenic culture medium respectively as demonstrated by the formation of alizarin-red staining matrix, alcian blue staining or Oil-Red-O staining intracellular droplets. There were no significant differences in cell growth rates between different cell types (data not shown).
3.2. Gene Expression of Putative Periodontal Markers in vitro.
3.2.1. Asporin
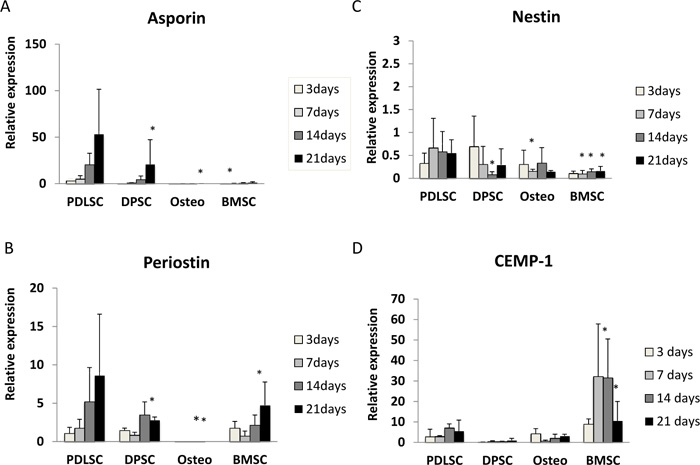
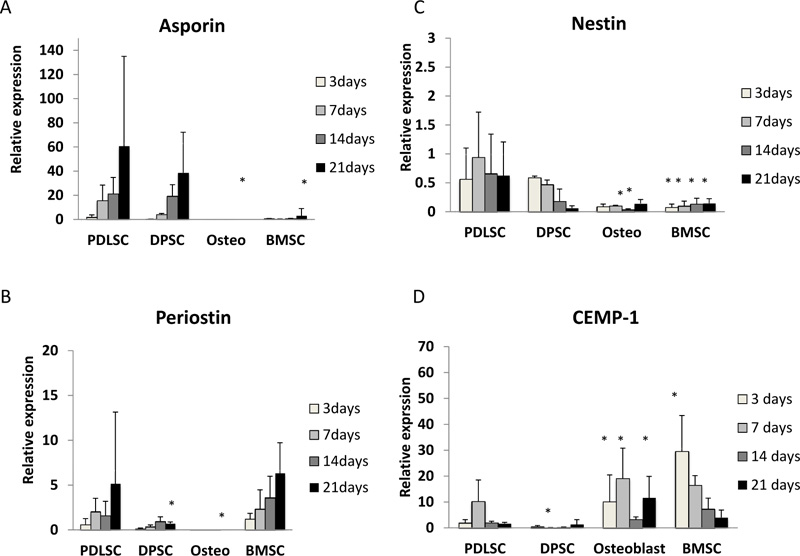
Asporin expression was seen mainly in PDLSC and, to a lesser extent, in DPSC. For cells grown in both normal and osteogenic medium, asporin expression in PDLSCs increased by around 50-fold over 21 days (Fig. 1A, 2A). DPSC also showed an increased expression of asporin with time but this was significantly less than in PDLSC at all timepoints. There were consistently low levels of expression of asporin by BMSCs and osteoblasts at all time points in either culture condition (Fig. 1A, 2A).
3.2.2. Periostin
Periostin expression was seen mainly in PDLSCs and BMSCs. In normal medium periostin expression was significantly increased in PDLSC by day 21, when compared to that seen in other cell types (Fig. 1B). In cultures with osteogenic medium, periostin expression in PDLSCs and BMSCs was similar and increased significantly with time in culture (Fig. 2B). There was no detectable expression of periostin in osteoblasts. In addition, periostin expression was lower in all cell types cultured in osteogenic media compared to those in normal media (Fig. 2B).
3.2.3. Nestin
Nestin expression was reduced at all time points when compared to the baseline (day 0). The expression of nestin was higher in PDLSCs and DPSCs when compared to BMSC and osteoblasts in both normal and osteogenic media (Fig. 1C, 2C).
3.2.4. CEMP-1
In normal medium, the highest levels of expression were seen in BMSCs, which reached its maximum at days 7 and 14 (Fig. 1D). In contrast expression of CEMP-1 was consistently low in PDLSC, DPSC and osteoblasts. In osteogenic medium, the greatest expression of CEMP-1 in BMSC was seen at day 3 (Fig. 2D) and declined at later time points. In addition, increased CEMP-1 expression was also seen in osteoblast cultures in osteogenic medium (Fig. 2D).
3.2.5. Expression of periodontal ligament associated proteins.
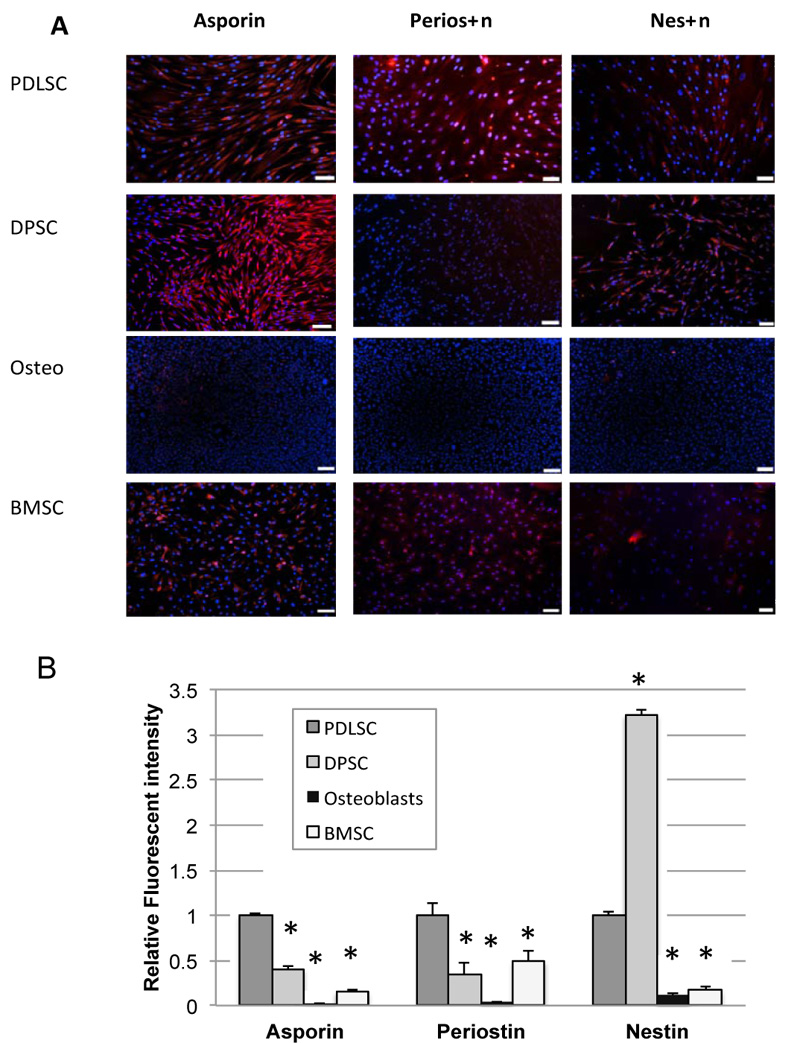
Immunofluorescence cytochemistry was used to test for protein expression. The results of immunofluorescence studies for asporin, periostin, and nestin are shown in Fig. (3). Positive staining for all 3 proteins was seen particularly in PDLSC, although nestin staining was also particularly high in DPSC. Immunofluorescence staining was not carried out for CEMP-1 as there was no significant expression of CEMP-1 seen in PDLSC at the mRNA level.
Quantification of integrated fluorescence intensity, which corrects for different numbers of cells between fields, is shown in Fig. (3B). Results demonstrate significantly higher expression of asporin, periostin, and nestin in PDLSC compared to BMSC. Staining in osteoblasts was almost non-existent throughout.
4. DISCUSSION
Phenotypic differences between MSCs derived from different tissues have been proposed as potentially of importance in selecting cell sources for optimal tissue engineering strategies. Some studies have tried to address this issue in dental stem cells and there is at least some evidence from non-human sources that there are quantitative (but not qualitative) differences in protein expression patterns between PDLSCs, BMSCs and DPSCs [18, 19].
We postulated that potential candidate genes to distinguish between PDLSC and other MSCs might reasonably be those of proteins that are highly expressed preferentially in the periodontal ligament, including asporin, periostin, nestin and cementum protein-1 (CEMP-1). This required that high expression levels of these proteins may occur constitutively in cells associated with the periodontal ligament, rather than be dependent on environmental cues present within the specific tissue itself.
The results of the studies described here firstly support the idea that asporin, periostin, and nestin are constitutively highly expressed in PDLSC, in line with previous studies demonstrating that they are abundant in the periodontal ligament. The 4th candidate marker that was investigated, CEMP-1, in contrast, was not associated with PDLSC but was constitutively expressed by BMSC. Interestingly, when cells were exposed to osteogenic medium, osteoblast cultures also showed CEMP-1 expression, but this was not seen with PDLSC or with DPSC. Although the studies carried out here were based largely on gene expression data, our studies of protein expression by immunofluorescence are largely in line with the findings at the mRNA level. However, we did not carry out a full series of experiments at later time points where some of the changes in gene expression are most evident, and quantification of immunofluorescence intensity is, at best, a semi-quantitative method of assessing protein expression.
The results suggest that these MSC lines isolated from different tissues show inherent differences in constitutive gene expression. PDLSC expressed high levels of asporin, periostin and nestin but not CEMP-1. In contrast, BMSC showed low levels of asporin and nestin expression but did express periostin and also expressed CEMP-1. DPSC expressed asporin and nestin, and to a lesser extent periostin, but not CEMP-1. Thus, each MSC cell type had a distinct pattern of expression of the candidate genes tested.
Ideally it would be helpful to test MSC cell phenotype differences in cells that were all derived from the same donor. However, unfortunately this was not feasible due to issues of tissue availability from donors. However, as we were able to test cells from 3 different donors for BMSC and PDLSC (and 2 from DPSC) and combine the data from these, our results show that differences in expression of these genes were more associated with differences in the source of tissue derivation than with differences between donors.
Although asporin expression is localised to the periodontal ligament, it has been shown to be expressed in a number of tissues, most notably in cartilage and bone, and is able to attach to collagen fibres and bind calcium [13]. Therefore, its expression in PDLSC was predicted, although the absence of expression in BMSCs, even after stimulation of osteogenic differentiation was perhaps less expected. Similarly, periostin has also been reported to be expressed in bone as well as in periodontal ligament. Thus, our results of periostin expression in both PDLSC and BMSC are consistent with these previous findings.
Nestin expression is considered a marker of neural crest-derived cells. Therefore, our results support these previous findings with high expression of nestin seen in the neural crest-derived PDLSC and DPSC, but not in BMSC or the osteoblast control cultures.
As already discussed, CEMP-1 expression has been described in the bone as well as in cementum. Thus, it was not unexpected to see CEMP-1 gene expression in BMSC, but much less expected that PDLSC did not express CEMP-1, even after 21 days of stimulation in osteogenic medium and despite the fact that these specific cell cultures are known to undergo osteoblastic differentiation and expression of osteoblast associated genes during culture in osteogenic medium as used here.
Overall the differences in expression patterns of these putative periodontal markers support the hypothesis of there being constitutive differences in mesenchymal stem cell populations derived from periodontal, dental pulp and bone marrow sources, which persist even following osteoblastic induction. The functional significance of these differences is currently uncertain. One clinically relevant issue is whether PDLSC may be particularly suited for application in periodontal regenerative tissue engineering procedures. Despite this possibility, mesenchymal stem cells from a range of different sources have all been reported to be able to contribute to the formation of new periodontal attachment in preclinical animal studies, and do not provide any evidence either way of the relative clinical effectiveness of MSC from difference sources.
CONCLUSION
There were clear differences in expression patterns between PDLSCs and BMSCs, which are suggestive of innate phenotypic differences between MSC types. PDLSC, in particular, expressed high levels of the matricellular proteins periostin and asporin, and the neural crest-derived cytoskeletal protein nestin. Gene expression of Cementum Protein-1 was mainly seen in BMSC and osteoblasts and not in PDLSC.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Ethical approval for obtaining the tissue samples was obtained from Kent NHS Research Ethics Committee, UK (Reference No: 11/LO/0259).
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures followed were in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written informed consent was obtained from each participant prior to the study.
AVAILABILITY OF DATA AND MATERIALS
The data that support the findings of this study are available from the corresponding author, [F.H], upon reasonable request.
FUNDING
DG is generously supported by the Directorate General of Higher Education Indonesia Ministry of Education. Grant number 373.33/E4.4/K/2012.
CONFLICTS OF INTEREST
The authors report no conflict of interest associated with this work.
ACKNOWLEDGEMENTS
Declared none.


